BitterDB is a free repository of bitter molecules. It contains information on compounds that were documented to taste bitter to humans or to activate bitter taste receptors in-vitro.
To cite BitterDB, please reference:
BitterDB: a database of bitter compounds.
Nucleic Acids Res. 2012 Wiener A, Shudler M, Levit A, Niv MY.
View on NAR website
Information about activation of human bitter taste receptors by bitter molecules can be found in BitterDB as described in the above paper, and for example by using this link: http://bitterdb.agri.huji.ac.il/bitterdb/dbbitter.php#receptorBrowse
We are grateful to the authors for allowing us to incorporate the molecular structrues of this database in ZINC.We assess the chemical diversity of a subset by clustering the molecules. First, we sort ligands by increasing molecular weight. Then, we use the SUBSET 1.0 algorithm ( Voigt JH, Bienfait B, Wang S, Nicklaus MC. JCICS, 2001, 41, 702-12) to progressively select compounds that differ from those previously selected by at least the Tanimoto cutoff, using ChemAxon default fingerprints. The resulting representatives have two interesting properties:
| Tanimoto Cutoff Level | 60% | 70% | 80% | 90% | 100% |
|---|---|---|---|---|---|
| Number of Representatives | 216 | 301 | 363 | 436 | 686 |
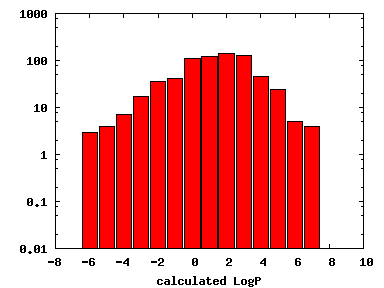
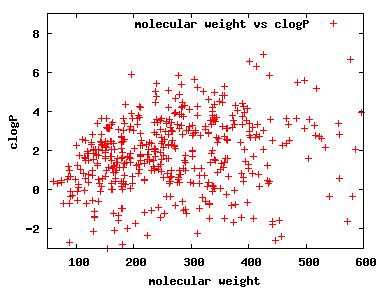
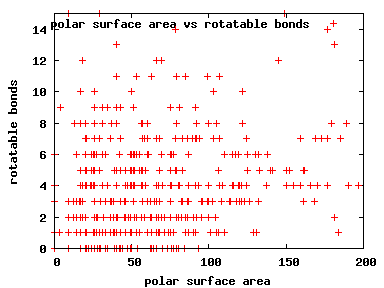
We compute the physical properties of each molecule in the subset, and graph them below.
Download Calculated Physical Properties



| Format | Reference(pH 7) | Mid(pH 6-8) | High(pH 8-9.5) | Low(pH 4.5-6) | Download Unix |
Download Windows |
|---|---|---|---|---|---|---|
| SMILES | All | All | All | All | ||
| MOL2 | All | All | All | All | Single Usual Metals All | Single Usual Metals All |
| SDF | All | All | All | All | Single Usual Metals All | Single Usual Metals All |
| Flexibase | Not Available | Not Available | Not Available | Not Available |