|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
2.92 |
-11.1 |
4 |
5 |
0 |
90 |
444.524 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR_HUMAN |
P04150
|
Glucocorticoid Receptor, Human |
0.1 |
0.38 |
Functional ≤ 10μM
|
|
Z100081 |
Z100081
|
PBMC (Peripheral Blood Mononuclear Cells) |
0.1 |
0.38 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
4.25 |
-22.44 |
1 |
6 |
0 |
93 |
538.584 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
11.17 |
-21.58 |
1 |
6 |
0 |
76 |
464.444 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM3-3-E |
Muscarinic Acetylcholine Receptor M3 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.1 |
0.42 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
0.1 |
0.42 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
0.1 |
0.42 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
0.199526231 |
0.41 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
0.158489319 |
0.42 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.1 |
0.42 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
0.1 |
0.42 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
0.1 |
0.42 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
0.199526231 |
0.41 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
0.158489319 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.59 |
14.06 |
-41.76 |
1 |
5 |
1 |
56 |
484.663 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.59 |
14.78 |
-62.29 |
0 |
5 |
0 |
59 |
483.655 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
-0.55 |
-12.57 |
3 |
7 |
0 |
92 |
482.821 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PSB8-1-E |
Proteasome Subunit Beta Type-8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
14 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
9.32 |
-14.74 |
4 |
12 |
0 |
158 |
719.924 |
20 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.94 |
12.61 |
-46.9 |
5 |
12 |
1 |
160 |
720.932 |
20 |
↓
|
|
|
|
Analogs
-
4533786
-
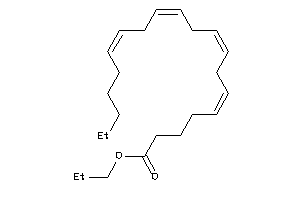
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
15.63 |
-5.46 |
0 |
2 |
0 |
26 |
330.512 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
JAK1-1-E |
Tyrosine-protein Kinase JAK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
JAK2-1-E |
Tyrosine-protein Kinase JAK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
JAK3-1-E |
Tyrosine-protein Kinase JAK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.39 |
Binding ≤ 10μM
|
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
9.25 |
-17.45 |
1 |
6 |
0 |
83 |
306.373 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF-1-E |
Cathepsin F (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
40 |
0.28 |
Binding ≤ 1μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
760 |
0.23 |
Binding ≤ 1μM
|
|
CATL2_HUMAN |
O60911
|
Cathepsin L2, Human |
75 |
0.27 |
Binding ≤ 1μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
120 |
0.26 |
Binding ≤ 1μM
|
|
CMA1_HUMAN |
P23946
|
Chymase, Human |
32 |
0.28 |
Binding ≤ 1μM
|
|
Z50643 |
Z50643
|
Hepatitis C Virus |
14 |
0.30 |
Binding ≤ 1μM
|
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
14 |
0.30 |
Binding ≤ 1μM
|
|
CATB_HUMAN |
P07858
|
Cathepsin B, Human |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF_HUMAN |
Q9UBX1
|
Cathepsin F, Human |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2_HUMAN |
O60911
|
Cathepsin L2, Human |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1_HUMAN |
P23946
|
Chymase, Human |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643 |
Z50643
|
Hepatitis C Virus |
14 |
0.30 |
Binding ≤ 10μM
|
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643 |
Z50643
|
Hepatitis C Virus |
200 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.18 |
-13.07 |
5 |
10 |
0 |
151 |
519.687 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF-1-E |
Cathepsin F (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
|
D2K2A8-1-V |
Hepatitis C Virus NS4A Protein (cluster #1 Of 1), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
40 |
0.28 |
Binding ≤ 1μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
760 |
0.23 |
Binding ≤ 1μM
|
|
CATL2_HUMAN |
O60911
|
Cathepsin L2, Human |
75 |
0.27 |
Binding ≤ 1μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
120 |
0.26 |
Binding ≤ 1μM
|
|
CMA1_HUMAN |
P23946
|
Chymase, Human |
32 |
0.28 |
Binding ≤ 1μM
|
|
Z50643 |
Z50643
|
Hepatitis C Virus |
14 |
0.30 |
Binding ≤ 1μM
|
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
14 |
0.30 |
Binding ≤ 1μM
|
|
D2K2A8_9HEPC |
D2K2A8
|
Hepatitis C Virus NS4A Protein, 9hepc |
14 |
0.30 |
Binding ≤ 1μM
|
|
CATB_HUMAN |
P07858
|
Cathepsin B, Human |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF_HUMAN |
Q9UBX1
|
Cathepsin F, Human |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2_HUMAN |
O60911
|
Cathepsin L2, Human |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1_HUMAN |
P23946
|
Chymase, Human |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643 |
Z50643
|
Hepatitis C Virus |
14 |
0.30 |
Binding ≤ 10μM
|
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
14 |
0.30 |
Binding ≤ 10μM
|
|
D2K2A8_9HEPC |
D2K2A8
|
Hepatitis C Virus NS4A Protein, 9hepc |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643 |
Z50643
|
Hepatitis C Virus |
200 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.12 |
-12.14 |
5 |
10 |
0 |
151 |
519.687 |
10 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
1998 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
8 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
3.57 |
-12.57 |
2 |
6 |
0 |
97 |
366.428 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SHH-1-E |
Sonic Hedgehog Protein (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
ALBU-1-E |
Serum Albumin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.27 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
7.07 |
-22.4 |
1 |
5 |
0 |
76 |
421.305 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.60 |
2.44 |
-15.62 |
1 |
3 |
0 |
42 |
139.154 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
9.62 |
-15.78 |
3 |
5 |
0 |
82 |
392.499 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
9.75 |
-11.2 |
2 |
5 |
0 |
76 |
452.538 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
3.98 |
-20.91 |
4 |
10 |
0 |
138 |
522.578 |
10 |
↓
|
|
|
|
Analogs
-
34636112
-
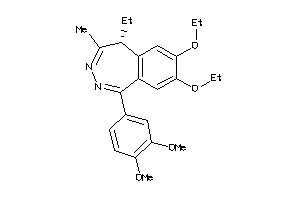
-
608106
-
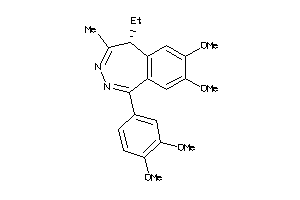
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
8.2 |
-14.87 |
0 |
6 |
0 |
62 |
382.46 |
6 |
↓
|
|
|
|
Analogs
-
3831552
-
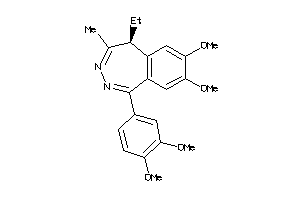
-
34636112
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
8.27 |
-14.96 |
0 |
6 |
0 |
62 |
382.46 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.66 |
3.68 |
-10.9 |
0 |
3 |
0 |
43 |
428.657 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.81 |
-0.84 |
-18.84 |
3 |
6 |
0 |
101 |
267.31 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.64 |
-1.63 |
-55.15 |
2 |
6 |
-1 |
100 |
266.302 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.81 |
-0.98 |
-62.25 |
4 |
6 |
1 |
103 |
268.318 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
3.01 |
-12.25 |
0 |
5 |
0 |
59 |
328.151 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
-0.93 |
-4.32 |
1 |
2 |
0 |
29 |
230.392 |
13 |
↓
|
|
|
|
Analogs
-
38141475
-
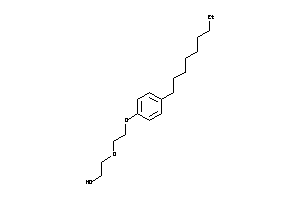
-
8214628
-
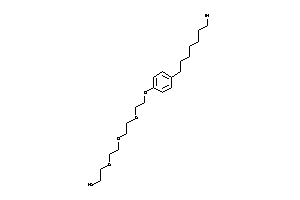
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
11.13 |
-18.22 |
1 |
10 |
0 |
103 |
616.833 |
35 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.42 |
-12.55 |
4 |
4 |
0 |
86 |
254.289 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
-0.02 |
-4.26 |
1 |
1 |
0 |
20 |
124.23 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.31 |
0.52 |
-43.59 |
1 |
1 |
-1 |
20 |
123.222 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.31 |
0.5 |
-43.08 |
1 |
1 |
-1 |
20 |
123.222 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
-0.07 |
-2.95 |
1 |
1 |
0 |
20 |
124.23 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.31 |
0.47 |
-38.82 |
1 |
1 |
-1 |
20 |
123.222 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
12.28 |
-21.69 |
1 |
7 |
0 |
107 |
508.558 |
8 |
↓
|
|
|
|
Analogs
-
6578890
-
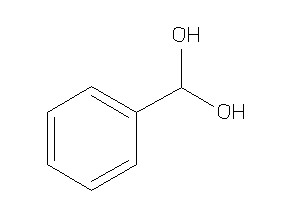
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
1.82 |
-4.35 |
1 |
1 |
0 |
20 |
108.14 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
7.55 |
-6.8 |
1 |
2 |
0 |
37 |
302.458 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
59 |
0.92 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1600 |
0.74 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.81 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.88 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.20 |
7.01 |
-33.25 |
0 |
3 |
1 |
26 |
160.237 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
59 |
0.92 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
1600 |
0.74 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.81 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
120 |
0.88 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.20 |
7.02 |
-33.28 |
0 |
3 |
1 |
26 |
160.237 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
329 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
-1.89 |
-17.12 |
2 |
9 |
0 |
111 |
459.506 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
0.53 |
0.72 |
Binding ≤ 1μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.1 |
0.78 |
Binding ≤ 1μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.1 |
0.78 |
Binding ≤ 1μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
0.53 |
0.72 |
Binding ≤ 1μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
0.53 |
0.72 |
Binding ≤ 1μM
|
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
0.53 |
0.72 |
Binding ≤ 10μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.1 |
0.78 |
Binding ≤ 10μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
0.53 |
0.72 |
Binding ≤ 10μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.1 |
0.78 |
Binding ≤ 10μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
0.53 |
0.72 |
Binding ≤ 10μM
|
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
10.1 |
0.62 |
Functional ≤ 10μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
10.1 |
0.62 |
Functional ≤ 10μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
10.1 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
-0.43 |
-12.34 |
1 |
3 |
0 |
38 |
243.306 |
4 |
↓
|
|
|
|
Analogs
-
3964523
-
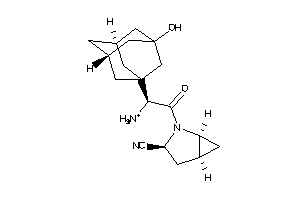
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
DPP8-1-E |
Dipeptidyl Peptidase VIII (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
240 |
0.40 |
Binding ≤ 10μM
|
|
DPP9-1-E |
Dipeptidyl Peptidase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
71 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
2.91 |
-12.9 |
3 |
5 |
0 |
90 |
315.417 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
7.05 |
-15.07 |
3 |
10 |
0 |
125 |
483.96 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.31 |
7.47 |
-37.87 |
4 |
10 |
1 |
127 |
484.968 |
9 |
↓
|
|
|
|
Analogs
-
1494180
-
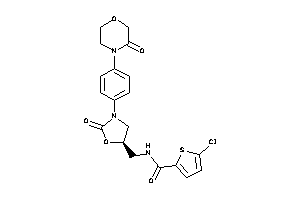
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
-3.62 |
-20.72 |
1 |
8 |
0 |
88 |
435.889 |
5 |
↓
|
|
|
|
Analogs
-
29562604
-
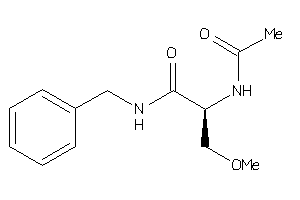
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
362 |
0.50 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3713 |
0.42 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1210 |
0.46 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
473 |
0.49 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
461 |
0.49 |
Binding ≤ 10μM
|
|
CAH2-14-E |
Carbonic Anhydrase II (cluster #14 Of 15), Eukaryotic |
Eukaryotes |
331 |
0.50 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
374 |
0.50 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
525 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
4565 |
0.42 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
341 |
0.50 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
412 |
0.50 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4446 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
353 |
0.50 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
461 |
0.49 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
362 |
0.50 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
331 |
0.50 |
Binding ≤ 1μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
374 |
0.50 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
525 |
0.49 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
353 |
0.50 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
341 |
0.50 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
412 |
0.50 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
473 |
0.49 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
461 |
0.49 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
362 |
0.50 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
331 |
0.50 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
374 |
0.50 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
525 |
0.49 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
353 |
0.50 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
4565 |
0.42 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
341 |
0.50 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
412 |
0.50 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
4446 |
0.42 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
3713 |
0.42 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
1210 |
0.46 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
473 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-2.84 |
-9.35 |
2 |
5 |
0 |
67 |
250.298 |
6 |
↓
|
|
|
|
Analogs
-
11617063
-
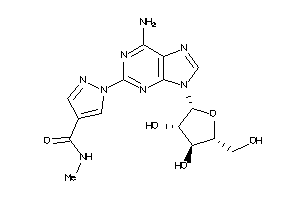
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
290 |
0.33 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
297 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.27 |
-3.31 |
-23.49 |
6 |
13 |
0 |
186 |
390.36 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.27 |
-2.93 |
-33.2 |
7 |
13 |
1 |
188 |
391.368 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.27 |
-7.78 |
-35.75 |
7 |
13 |
1 |
188 |
391.368 |
4 |
↓
|
|
|
|
Analogs
-
1490477
-
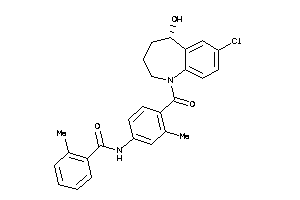
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
V1AR-1-E |
Vasopressin V1a Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
V2R-1-E |
Vasopressin V2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
-0.59 |
-11.63 |
2 |
5 |
0 |
69 |
448.95 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
11.25 |
-10.79 |
0 |
4 |
0 |
47 |
373.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
11.7 |
-15.15 |
0 |
4 |
0 |
47 |
373.449 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.74 |
1.5 |
-10.16 |
2 |
5 |
0 |
74 |
238.197 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1_HUMAN |
P21554
|
Cannabinoid CB1 Receptor, Human |
0.19 |
0.45 |
Binding ≤ 1μM
|
|
CNR1_RAT |
P20272
|
Cannabinoid CB1 Receptor, Rat |
0.17 |
0.46 |
Binding ≤ 1μM
|
|
CNR1_MOUSE |
P47746
|
Cannabinoid CB1 Receptor, Mouse |
1.8 |
0.41 |
Binding ≤ 1μM
|
|
CNR2_MOUSE |
P47936
|
Cannabinoid CB2 Receptor, Mouse |
514 |
0.29 |
Binding ≤ 1μM
|
|
CNR2_HUMAN |
P34972
|
Cannabinoid CB2 Receptor, Human |
1.9 |
0.41 |
Binding ≤ 1μM
|
|
CNR1_MOUSE |
P47746
|
Cannabinoid CB1 Receptor, Mouse |
1.8 |
0.41 |
Binding ≤ 10μM
|
|
CNR1_HUMAN |
P21554
|
Cannabinoid CB1 Receptor, Human |
0.19 |
0.45 |
Binding ≤ 10μM
|
|
CNR1_RAT |
P20272
|
Cannabinoid CB1 Receptor, Rat |
0.17 |
0.46 |
Binding ≤ 10μM
|
|
CNR2_MOUSE |
P47936
|
Cannabinoid CB2 Receptor, Mouse |
1640 |
0.27 |
Binding ≤ 10μM
|
|
CNR2_HUMAN |
P34972
|
Cannabinoid CB2 Receptor, Human |
1.9 |
0.41 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
2790 |
0.26 |
Binding ≤ 10μM |
|
CNR1_MOUSE |
P47746
|
Cannabinoid CB1 Receptor, Mouse |
8990 |
0.24 |
Functional ≤ 10μM
|
|
CNR1_HUMAN |
P21554
|
Cannabinoid CB1 Receptor, Human |
0.11 |
0.46 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
0.25 |
0.45 |
Functional ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
1.98 |
0.41 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
1.1 |
0.42 |
Functional ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
5.3 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.22 |
12.29 |
-7.95 |
1 |
5 |
0 |
50 |
463.796 |
4 |
↓
|
|