|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(NE,4R)-N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2-ylidene)-4-[(3R,5S,7R,8S,9S,10S,12S,13R,14S,17R)-
(NE,4R)-N-(3-methyl-5-sulfamoyl-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
700 |
0.22 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
12 |
0.28 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
115 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.08 |
-0.13 |
-75.09 |
6 |
10 |
1 |
165 |
585.813 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.08 |
-0.27 |
-46.76 |
5 |
10 |
0 |
164 |
584.805 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(NE,4R)-N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2-ylidene)-4-[(3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-
(NE,4R)-N-(3-methyl-5-sulfamoyl-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
700 |
0.22 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
12 |
0.28 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
115 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.08 |
0.03 |
-75.47 |
6 |
10 |
1 |
165 |
585.813 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.08 |
-0.11 |
-46.96 |
5 |
10 |
0 |
164 |
584.805 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10S,13R,14S,17R)-14-hydroxy-3-[(2R,4R,5S,6R)-5-hydroxy-4-methoxy-6-methyl-tetrahydro
3-[(3S,5S,8R,9S,10S,13R,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
1800 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
9.39 |
-13.83 |
2 |
6 |
0 |
85 |
516.719 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S,8R,9S,10S,13S,14S,17R)-3-hydroxy-13-methyl-1,2,3,4,5,6,7,8,9,10,11,12,14,15,16,17-hexadecahyd
(3R,5S,8R,9S,10S,13S,14S,17R)-3-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
7.9 |
-8.62 |
1 |
2 |
0 |
44 |
287.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S,8S,9S,10S,13S,14S,17R)-3-hydroxy-13-methyl-1,2,3,4,5,6,7,8,9,10,11,12,14,15,16,17-hexadecahyd
(3R,5S,8S,9S,10S,13S,14S,17R)-3-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
7.76 |
-9.87 |
1 |
2 |
0 |
44 |
287.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5S,8S,9R,10S,13S,14S,17R)-3-hydroxy-13-methyl-1,2,3,4,5,6,7,8,9,10,11,12,14,15,16,17-hexadecahyd
(3R,5S,8S,9R,10S,13S,14S,17R)-3-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
7.38 |
-9.71 |
1 |
2 |
0 |
44 |
287.447 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
3814431
-
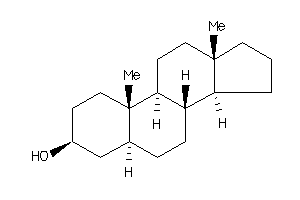
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R,8S,9R,10R,13S,14R,17R)-17-[(1R)-1,5-dimethylhexyl]-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15
(3R,5R,8S,9R,10R,13S,14R,17R)-17…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.80 |
12.45 |
-1.36 |
1 |
1 |
0 |
20 |
388.68 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
4.51 |
-9.9 |
3 |
4 |
0 |
78 |
388.548 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3R,5S,8S,9R,10S,13S,14S,17R)-3-hydroxy-3,10,13-trimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetra
1-[(3R,5S,8S,9R,10S,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
8.85 |
-6.86 |
1 |
2 |
0 |
37 |
332.528 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3R,5S,8S,9R,10S,13S,14R,17R)-3-hydroxy-3,10,13-trimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetra
1-[(3R,5S,8S,9R,10S,13S,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
8.33 |
-5.12 |
1 |
2 |
0 |
37 |
332.528 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,5S)-3-[(3S,5R,8S,9S,10R,13S,14R,17S)-3-[(3,5-difluorophenyl)methylamino]-4,4,8,10-tetramethyl-1,
(3S,5S)-3-[(3S,5R,8S,9S,10R,13S,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.27 |
15.85 |
-52.38 |
3 |
4 |
1 |
63 |
584.812 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.27 |
14.82 |
-7.79 |
2 |
4 |
0 |
59 |
583.804 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,5S)-3-[(3R,5R,8S,9S,10R,13S,14R,17S)-3-[(3,5-difluorophenyl)methylamino]-4,4,8,10-tetramethyl-1,
(3S,5S)-3-[(3R,5R,8S,9S,10R,13S,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.27 |
15.88 |
-52.78 |
3 |
4 |
1 |
63 |
584.812 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
7.27 |
15.18 |
-7.66 |
2 |
4 |
0 |
59 |
583.804 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-5-(3-aminoanilino)-2-[[(4S)-4-[(3S,5R,7R,8S,9R,10S,13R,14S,17S)-3,7-dihydroxy-10,13-dimethyl-2,
(2S)-5-(3-aminoanilino)-2-[[(4S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.43 |
-69.29 |
6 |
9 |
-1 |
165 |
610.816 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S)-5-(3-aminoanilino)-2-[[(4R)-4-[(3S,5R,7R,8S,9R,10S,13R,14S,17S)-3,7-dihydroxy-10,13-dimethyl-2,
(2S)-5-(3-aminoanilino)-2-[[(4R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.57 |
-63.88 |
6 |
9 |
-1 |
165 |
610.816 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-5-(3-aminoanilino)-2-[[(4S)-4-[(3S,5R,7R,8S,9R,10S,13R,14S,17S)-3,7-dihydroxy-10,13-dimethyl-2,
(2R)-5-(3-aminoanilino)-2-[[(4S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.5 |
-68.43 |
6 |
9 |
-1 |
165 |
610.816 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R)-5-(3-aminoanilino)-2-[[(4R)-4-[(3S,5R,7R,8S,9R,10S,13R,14S,17S)-3,7-dihydroxy-10,13-dimethyl-2,
(2R)-5-(3-aminoanilino)-2-[[(4R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
7.62 |
-64.47 |
6 |
9 |
-1 |
165 |
610.816 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R,8R,9R,10R,13R,14S,17S)-14-hydroxy-13-methyl-17-(5-oxo-2H-furan-3-yl)-3-[(2R,3S,4S,5R,6R)-3,4,
(3R,5R,8R,9R,10R,13R,14S,17S)-14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
2.99 |
-20.4 |
4 |
9 |
0 |
143 |
534.646 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R,8R,9R,10R,13R,14S,17S)-14-hydroxy-13-methyl-17-(5-oxo-2H-furan-3-yl)-3-[(2R,3S,4S,5R,6S)-3,4,
(3R,5R,8R,9R,10R,13R,14S,17S)-14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
2.5 |
-17.87 |
4 |
9 |
0 |
143 |
534.646 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R,8R,9R,10R,13R,14S,17S)-14-hydroxy-13-methyl-17-(5-oxo-2H-furan-3-yl)-3-[(2R,3S,4S,5S,6R)-3,4,
(3R,5R,8R,9R,10R,13R,14S,17S)-14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.84 |
-19.05 |
4 |
9 |
0 |
143 |
534.646 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,5R,8R,9R,10R,13R,14S,17S)-14-hydroxy-13-methyl-17-(5-oxo-2H-furan-3-yl)-3-[(2R,3S,4S,5S,6S)-3,4,
(3R,5R,8R,9R,10R,13R,14S,17S)-14…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.49 |
-18.85 |
4 |
9 |
0 |
143 |
534.646 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-4-[(3R,5S,7R,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16
(4R)-4-[(3R,5S,7R,8R,9S,10S,13R,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
7.89 |
-44.82 |
4 |
6 |
1 |
77 |
490.753 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.00 |
5.54 |
-11.84 |
3 |
6 |
0 |
76 |
489.745 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-N-(4-methylpiperazin-1-yl)-4-[(3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-3,7,12-trihydroxy-10,13-dime
(4R)-N-(4-methylpiperazin-1-yl)-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
4.74 |
-47.42 |
5 |
7 |
1 |
97 |
506.752 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.09 |
2.38 |
-14.55 |
4 |
7 |
0 |
96 |
505.744 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4R)-4-[(3R,5S,7S,8R,9S,10S,13R,14S,17R)-3,7-dihydroxy-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16
(4R)-4-[(3R,5S,7S,8R,9S,10S,13R,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
7.45 |
-44.24 |
4 |
6 |
1 |
77 |
490.753 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.00 |
5.09 |
-11.69 |
3 |
6 |
0 |
76 |
489.745 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1S)-1-hydroxyethyl]-10,13-dimethyl-1,2,3,4,5,6,7,8,
1-[(3S,5S,8S,9R,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
6.73 |
-6.59 |
2 |
3 |
0 |
58 |
362.554 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1R)-1-hydroxyethyl]-10,13-dimethyl-1,2,3,4,5,6,7,8,
1-[(3S,5S,8S,9R,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
8.4 |
-5.97 |
2 |
3 |
0 |
58 |
362.554 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1S)-1-hydroxyethyl]-10,13-dimethyl-1,2,3,4,5,6,7,8,
1-[(3S,5S,8S,9R,10S,13R,14R,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
6 |
-6.91 |
2 |
3 |
0 |
58 |
362.554 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1R)-1-hydroxyethyl]-10,13-dimethyl-1,2,3,4,5,6,7,8,
1-[(3S,5S,8S,9R,10S,13R,14R,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
6.42 |
-6.41 |
2 |
3 |
0 |
58 |
362.554 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1S)-1-hydroxyethyl]-10,13-dimethyl-1
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-0.39 |
-10.74 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1R)-1-hydroxyethyl]-10,13-dimethyl-1
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
1.04 |
-9.52 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1S)-1-hydroxyethyl]-10,13-dimethyl-1
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-0.14 |
-10.25 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1R)-1-hydroxyethyl]-10,13-dimethyl-1
(2S,3S,4S,6S)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
400 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-0.69 |
-11.98 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
-
42764263
-
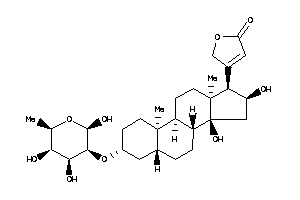
-
42764268
-
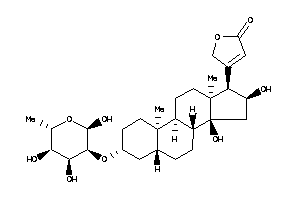
-
42764273
-
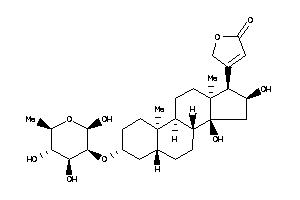
-
42764276
-
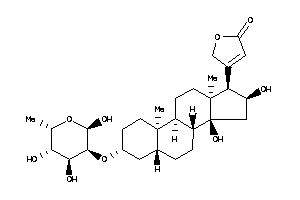
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10R,13S,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
3-[(3S,5S,8R,9S,10R,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.77 |
2.13 |
-16.02 |
5 |
9 |
0 |
146 |
536.662 |
4 |
↓
|
|
|
|
Analogs
-
42764263
-

-
42764268
-

-
42764273
-

-
42764276
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10S,13S,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
3-[(3S,5S,8R,9S,10S,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.77 |
1.97 |
-17.71 |
5 |
9 |
0 |
146 |
536.662 |
4 |
↓
|
|
|
|
Analogs
-
42764263
-

-
42764268
-

-
42764273
-

-
42764276
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3R,5S,8R,9S,10R,13S,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
3-[(3R,5S,8R,9S,10R,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.77 |
1.79 |
-16.75 |
5 |
9 |
0 |
146 |
536.662 |
4 |
↓
|
|
|
|
Analogs
-
42764263
-

-
42764268
-

-
42764273
-

-
42764276
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3R,5S,8R,9S,10S,13S,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
3-[(3R,5S,8R,9S,10S,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
3 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.77 |
1.49 |
-16.7 |
5 |
9 |
0 |
146 |
536.662 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-4,5-dihydroxy-6-[[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1S)-
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-3.54 |
-15.24 |
7 |
11 |
0 |
179 |
628.8 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-4,5-dihydroxy-6-[[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1R)-
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-2.11 |
-13.46 |
7 |
11 |
0 |
179 |
628.8 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-4,5-dihydroxy-6-[[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1S)-
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-3.3 |
-14.16 |
7 |
11 |
0 |
179 |
628.8 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-4,5-dihydroxy-6-[[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1R)-
(2R,4R,5R,6R)-6-[[(2R,4R,5R,6R)-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-3.85 |
-16.16 |
7 |
11 |
0 |
179 |
628.8 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
1-[(3S,5S,8S,9R,10S,13R,14R,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
1.72 |
-14.17 |
5 |
8 |
0 |
137 |
496.641 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
1-[(3S,5S,8S,9R,10S,13R,14R,17S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
0.33 |
-12.48 |
5 |
8 |
0 |
137 |
496.641 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
1-[(3S,5S,8S,9R,10S,13R,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
0.04 |
-11.14 |
5 |
8 |
0 |
137 |
496.641 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-[(3S,5S,8S,9R,10S,13R,14S,17S)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4S,5S,6R)-3,4,5,6-tetrahydroxy
1-[(3S,5S,8S,9R,10S,13R,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
0.28 |
-14.08 |
5 |
8 |
0 |
137 |
496.641 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1S)-1-hydroxyethyl]-10,13-dimethyl-1
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-0.27 |
-11.04 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,10S,13R,14R,17R)-14-hydroxy-17-[(1R)-1-hydroxyethyl]-10,13-dimethyl-1
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
1.17 |
-9.14 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1S)-1-hydroxyethyl]-10,13-dimethyl-1
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-0.03 |
-9.87 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,10S,13R,14R,17S)-14-hydroxy-17-[(1R)-1-hydroxyethyl]-10,13-dimethyl-1
(2R,4R,5R,6R)-6-[[(3S,5S,8S,9R,1…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
650 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-0.57 |
-12.01 |
5 |
7 |
0 |
120 |
482.658 |
4 |
↓
|
|
|
|
Analogs
-
31879449
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10R,13S,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4R,6R)-3,4,6-trihydroxytetrahy
3-[(3S,5S,8R,9S,10R,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
3.55 |
-15.32 |
4 |
8 |
0 |
126 |
520.663 |
4 |
↓
|
|
|
|
Analogs
-
31879449
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10S,13S,14S,17R)-14-hydroxy-10,13-dimethyl-3-[[(2R,3R,4R,6R)-3,4,6-trihydroxytetrahy
3-[(3S,5S,8R,9S,10S,13S,14S,17R)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
9 |
0.30 |
Binding ≤ 1μM
|
|
AT1A1_HUMAN |
P05023
|
Sodium/potassium-transporting ATPase Alpha-1 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A2_HUMAN |
P50993
|
Sodium/potassium-transporting ATPase Alpha-2 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A3_HUMAN |
P13637
|
Sodium/potassium-transporting ATPase Alpha-3 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1A4_HUMAN |
Q13733
|
Sodium/potassium-transporting ATPase Alpha-4 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1_PIG |
P05027
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Pig |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B1_HUMAN |
P05026
|
Sodium/potassium-transporting ATPase Beta-1 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B2_HUMAN |
P14415
|
Sodium/potassium-transporting ATPase Beta-2 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
AT1B3_HUMAN |
P54709
|
Sodium/potassium-transporting ATPase Beta-3 Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
|
ATNG_HUMAN |
P54710
|
Sodium/potassium-transporting ATPase Gamma Chain, Human |
9 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
3.4 |
-17.15 |
4 |
8 |
0 |
126 |
520.663 |
4 |
↓
|
|