|
|
Analogs
-
402782
-
-
4095869
-
-
4095870
-
-
4095871
-
-
4095872
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
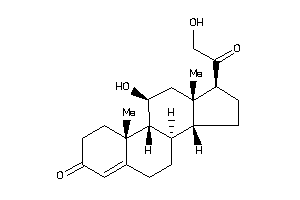
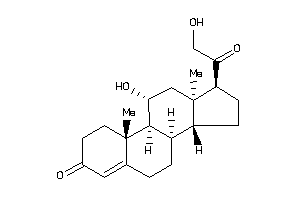
(8R,9R,10S,11R,13S,14S,17R)-17-glycoloyl-11-hydroxy-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dod
(8R,9R,10S,11R,13S,14S,17R)-17-g…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
-1.71 |
-10.96 |
2 |
4 |
0 |
74 |
346.467 |
2 |
↓
|
|
|
|
Analogs
-
43366257
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
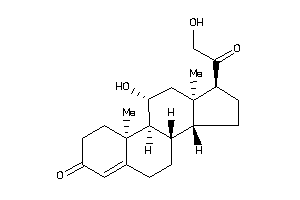
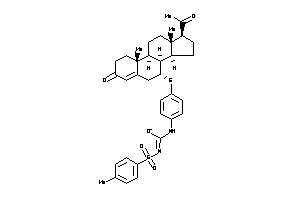
3-[4-[[(7R,8R,9R,10R,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodec
3-[4-[[(7R,8R,9R,10R,13S,14S,17S…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
12.4 |
-70.27 |
1 |
7 |
-1 |
116 |
633.856 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.07 |
12.09 |
-29.32 |
2 |
7 |
0 |
113 |
634.864 |
7 |
↓
|
|
|
|
Analogs
-
43366257
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
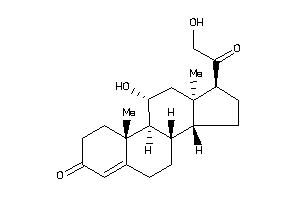
3-[4-[[(7R,8S,9R,10R,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodec
3-[4-[[(7R,8S,9R,10R,13S,14S,17S…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.07 |
12.53 |
-61.71 |
1 |
7 |
-1 |
116 |
633.856 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.07 |
12.28 |
-26.46 |
2 |
7 |
0 |
113 |
634.864 |
7 |
↓
|
|
|
|
Analogs
-
44201696
-
-
44201698
-
-
3965140
-
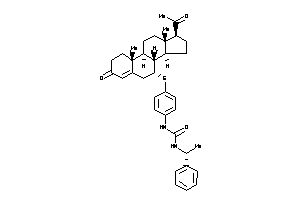
Draw
Identity
99%
90%
80%
70%
Popular Name:
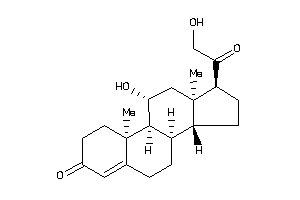
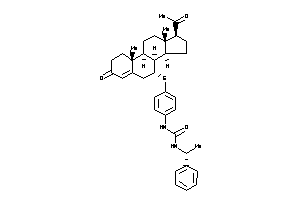
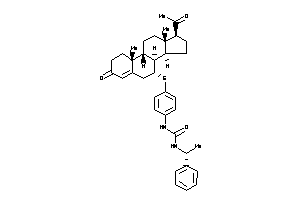
3-[4-[[(7R,8S,9R,10R,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodec
3-[4-[[(7R,8S,9R,10R,13S,14S,17S…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81008-1-O |
Human Breast Cancer Cell Lines (cluster #1 Of 1), Other |
Other |
800 |
0.20 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81008 |
Z81008
|
Human Breast Cancer Cell Lines |
600 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.05 |
16.58 |
-22.98 |
2 |
5 |
0 |
75 |
584.826 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14S,17S)-17-acetyl-13,17-dimethyl-1,2,6,7,8,9,10,11,12,14,15,16-dodecahydrocyclopenta
(8S,9R,10R,13S,14S,17S)-17-acety…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.57 |
-10.83 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(7R,8S,9S,10R,13S,14S,17S)-17-hydroxy-7,13,17-trimethyl-1,2,6,7,8,9,10,11,12,14,15,16-dodecahydrocyc
(7R,8S,9S,10R,13S,14S,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
7.8 |
-8.16 |
1 |
2 |
0 |
37 |
302.458 |
0 |
↓
|
|
|
|
Analogs
-
6778072
-
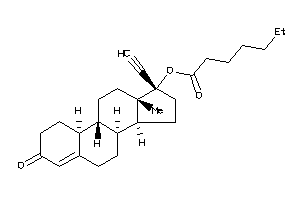
-
6778073
-
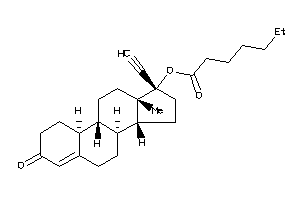
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.37 |
15.51 |
-11.96 |
0 |
3 |
0 |
43 |
410.598 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13S,14S,17S)-N-[1-(4-chlorophenyl)cyclopentyl]-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,
(8S,9S,10R,13S,14S,17S)-N-[1-(4-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.11 |
15.03 |
-11.75 |
1 |
3 |
0 |
46 |
494.119 |
3 |
↓
|
|
|
|
Analogs
-
35498275
-
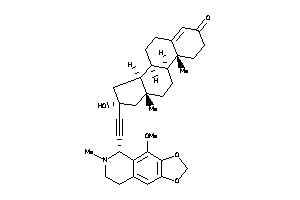
-
35498278
-
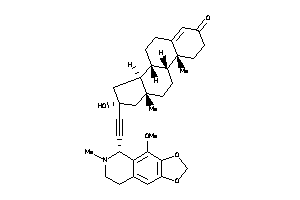
-
35498281
-
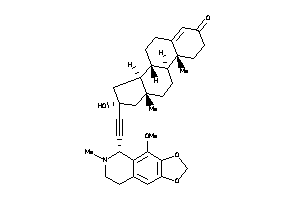
-
36512120
-
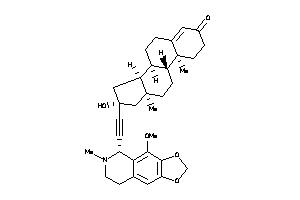
-
36512122
-
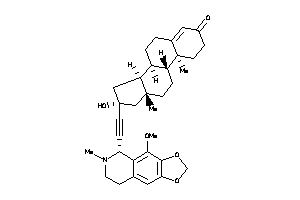
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13R,14S,16R)-16-hydroxy-16-[2-[(5R)-4-methoxy-6-methyl-7,8-dihydro-5H-[1,3]dioxolo[4,5-g]
(8R,9R,10R,13R,14S,16R)-16-hydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
10.04 |
-13.71 |
1 |
6 |
0 |
68 |
531.693 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.37 |
12.38 |
-49.9 |
2 |
6 |
1 |
69 |
532.701 |
1 |
↓
|
|
|
|
Analogs
-
35498278
-

-
35498281
-

-
36512120
-

-
36512122
-

-
36512124
-
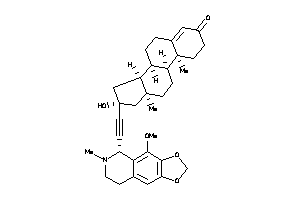
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9S,10R,13R,14S,16R)-16-hydroxy-16-[2-[(5R)-4-methoxy-6-methyl-7,8-dihydro-5H-[1,3]dioxolo[4,5-g]
(8R,9S,10R,13R,14S,16R)-16-hydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
9.96 |
-13.92 |
1 |
6 |
0 |
68 |
531.693 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.37 |
12.31 |
-53.71 |
2 |
6 |
1 |
69 |
532.701 |
1 |
↓
|
|
|
|
Analogs
-
35498281
-

-
36512120
-

-
36512122
-

-
36512124
-

-
36512126
-
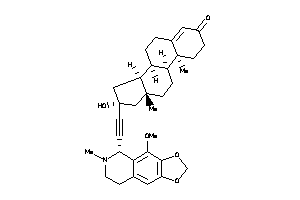
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13R,14S,16R)-16-hydroxy-16-[2-[(5R)-4-methoxy-6-methyl-7,8-dihydro-5H-[1,3]dioxolo[4,5-g]
(8S,9R,10R,13R,14S,16R)-16-hydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
10.09 |
-16.06 |
1 |
6 |
0 |
68 |
531.693 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.37 |
12.42 |
-58.19 |
2 |
6 |
1 |
69 |
532.701 |
1 |
↓
|
|
|
|
Analogs
-
36512120
-

-
36512122
-

-
36512124
-

-
36512126
-

-
35498272
-
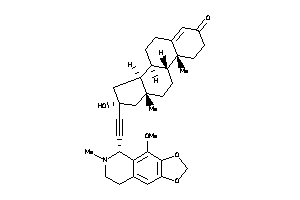
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13R,14S,16R)-16-hydroxy-16-[2-[(5R)-4-methoxy-6-methyl-7,8-dihydro-5H-[1,3]dioxolo[4,5-g]
(8S,9S,10R,13R,14S,16R)-16-hydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.37 |
9.78 |
-15.8 |
1 |
6 |
0 |
68 |
531.693 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.37 |
12.1 |
-58.32 |
2 |
6 |
1 |
69 |
532.701 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13R,14R)-13-[(1R)-1-hydroxyprop-2-ynyl]-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyc
(8R,9R,10R,13R,14R)-13-[(1R)-1-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
7.91 |
-8.19 |
1 |
2 |
0 |
37 |
298.426 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13S,14R)-13-[(1R)-1-hydroxyprop-2-ynyl]-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyc
(8R,9R,10R,13S,14R)-13-[(1R)-1-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
8.57 |
-8.11 |
1 |
2 |
0 |
37 |
298.426 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13R,14S)-13-[(1R)-1-hydroxyprop-2-ynyl]-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyc
(8R,9R,10R,13R,14S)-13-[(1R)-1-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
7.89 |
-6.95 |
1 |
2 |
0 |
37 |
298.426 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13S,14S)-13-[(1R)-1-hydroxyprop-2-ynyl]-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyc
(8R,9R,10R,13S,14S)-13-[(1R)-1-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
7.91 |
-8.27 |
1 |
2 |
0 |
37 |
298.426 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R)-2-oxo-1-[(8R,9S,10R,13R,14S)-3-oxo-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]
[(1R)-2-oxo-1-[(8R,9S,10R,13R,14…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5100 |
0.28 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5100 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
11.98 |
-13.28 |
0 |
4 |
0 |
60 |
358.478 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S)-2-oxo-1-[(8R,9S,10R,13R,14S)-3-oxo-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]
[(1S)-2-oxo-1-[(8R,9S,10R,13R,14…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5100 |
0.28 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5100 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
11.62 |
-11.23 |
0 |
4 |
0 |
60 |
358.478 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13R,14R)-13-[(1S)-1-hydroxyprop-2-ynyl]-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyc
(8R,9R,10R,13R,14R)-13-[(1S)-1-h…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
160 |
0.43 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
160 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
7.81 |
-7.75 |
1 |
2 |
0 |
37 |
298.426 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9R,10R,13S,14R)-13-[(1S)-1-hydroxyprop-2-ynyl]-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyc
(8R,9R,10R,13S,14R)-13-[(1S)-1-h…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
160 |
0.43 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
160 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
7.63 |
-8.26 |
1 |
2 |
0 |
37 |
298.426 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9S,10R,13S,14S)-13-acetonyl-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthr
(8R,9S,10R,13S,14S)-13-acetonyl-…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.42 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
10.69 |
-11.24 |
0 |
2 |
0 |
34 |
300.442 |
2 |
↓
|
|
|
|
Analogs
-
11990702
-
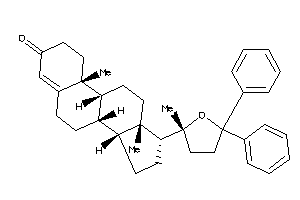
-
11990703
-
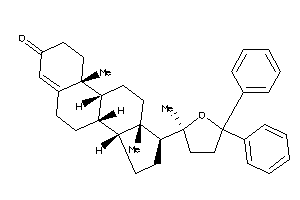
-
11990704
-
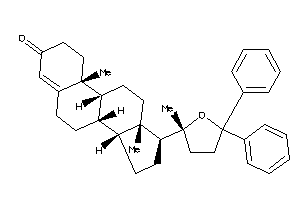
-
36646313
-
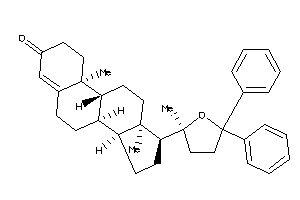
-
36646314
-
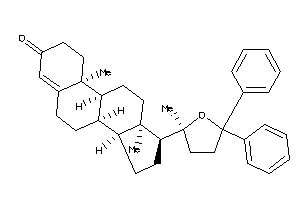
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14R,17R)-10,13-dimethyl-17-[(2S)-2-methyl-5,5-diphenyl-tetrahydrofuran-2-yl]-1,2,6,7,
(8S,9R,10R,13S,14R,17R)-10,13-di…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.71 |
3.06 |
-7.71 |
0 |
2 |
0 |
26 |
508.746 |
3 |
↓
|
|
|
|
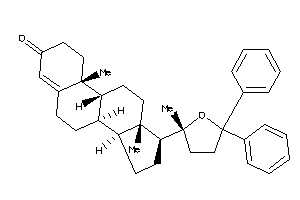
Analogs
-
11990703
-

-
11990704
-

-
36646313
-

-
36646314
-

-
39252940
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14R,17R)-10,13-dimethyl-17-[(2R)-2-methyl-5,5-diphenyl-tetrahydrofuran-2-yl]-1,2,6,7,
(8S,9R,10R,13S,14R,17R)-10,13-di…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.71 |
3.82 |
-6.33 |
0 |
2 |
0 |
26 |
508.746 |
3 |
↓
|
|
|
|
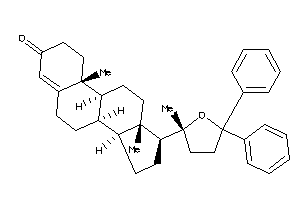
Analogs
-
11990704
-

-
36646313
-

-
36646314
-

-
39252940
-

-
39252941
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14R,17S)-10,13-dimethyl-17-[(2S)-2-methyl-5,5-diphenyl-tetrahydrofuran-2-yl]-1,2,6,7,
(8S,9R,10R,13S,14R,17S)-10,13-di…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.71 |
3.97 |
-6.96 |
0 |
2 |
0 |
26 |
508.746 |
3 |
↓
|
|
|
|
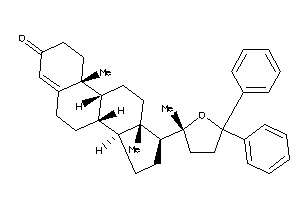
Analogs
-
36646313
-

-
36646314
-

-
39252940
-

-
39252941
-

-
39252942
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10R,13S,14R,17S)-10,13-dimethyl-17-[(2R)-2-methyl-5,5-diphenyl-tetrahydrofuran-2-yl]-1,2,6,7,
(8S,9R,10R,13S,14R,17S)-10,13-di…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.71 |
3.11 |
-9.74 |
0 |
2 |
0 |
26 |
508.746 |
3 |
↓
|
|
|
|
Analogs
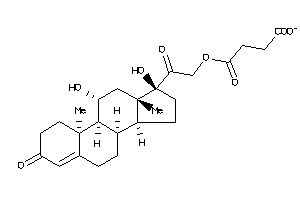
-
2123273
-
-
3872844
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
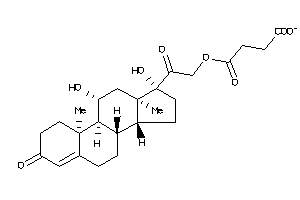
4-[2-[(8R,9S,10R,11S,13S,14S,17R)-11,17-dihydroxy-10,13-dimethyl-3-oxo-2,6,7,8,9,11,12,14,15,16-deca
4-[2-[(8R,9S,10R,11S,13S,14S,17R…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
8.07 |
-50.15 |
2 |
8 |
-1 |
141 |
461.531 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
33865172
-
-
34475380
-
-
34475381
-
-
40848053
-
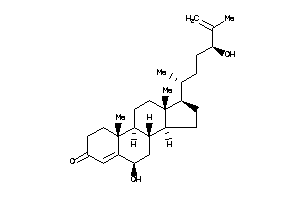
-
40848054
-
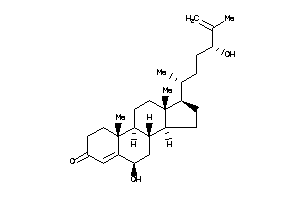
Draw
Identity
99%
90%
80%
70%
Popular Name:
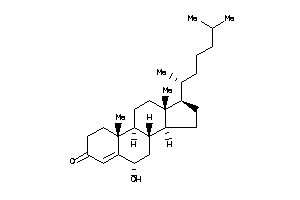
(6R,8S,9S,10R,13R,14R,17R)-17-[(1S)-1,5-dimethylhexyl]-6-hydroxy-10,13-dimethyl-1,2,6,7,8,9,11,12,14
(6R,8S,9S,10R,13R,14R,17R)-17-[(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.52 |
12.06 |
-5.05 |
1 |
2 |
0 |
37 |
400.647 |
5 |
↓
|
|
|
|
Analogs
-
33865172
-

-
34475380
-

-
34475381
-

-
40848053
-

-
40848054
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
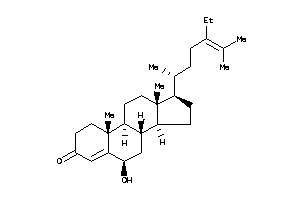
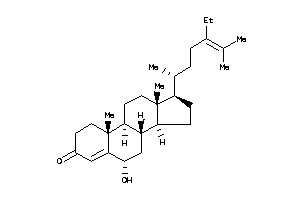
(6R,8S,9S,10R,13R,14S,17R)-17-[(1S)-1,5-dimethylhexyl]-6-hydroxy-10,13-dimethyl-1,2,6,7,8,9,11,12,14
(6R,8S,9S,10R,13R,14S,17R)-17-[(…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.52 |
12.05 |
-5.28 |
1 |
2 |
0 |
37 |
400.647 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9S,10R,13R,14R,17R)-17-[(1S)-1,5-dimethylhexyl]-10,13-dimethyl-1,2,6,7,8,9,11,12,14,15,16,17-dod
(8S,9S,10R,13R,14R,17R)-17-[(1S)…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.44 |
15.07 |
-5.48 |
0 |
1 |
0 |
17 |
384.648 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8R,9R,10R,13S,14R,17S)-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]
[(8R,9R,10R,13S,14R,17S)-10,13-d…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
10.89 |
-11.05 |
0 |
3 |
0 |
43 |
330.468 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8R,9R,10R,13S,14S,17S)-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]
[(8R,9R,10R,13S,14S,17S)-10,13-d…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
11.29 |
-11.19 |
0 |
3 |
0 |
43 |
330.468 |
2 |
↓
|
|
|
|
Analogs
-
16968900
-
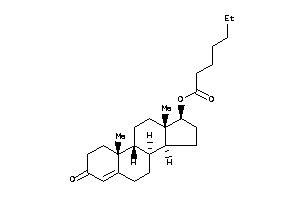
-
38613267
-
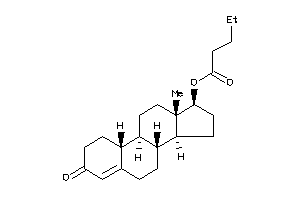
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8R,9R,10R,13S,14R,17S)-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]
[(8R,9R,10R,13S,14R,17S)-10,13-d…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
14.04 |
-11.91 |
0 |
3 |
0 |
43 |
386.576 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8R,9R,10R,13S,14S,17S)-10,13-dimethyl-3-oxo-1,2,6,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta[a]
[(8R,9R,10R,13S,14S,17S)-10,13-d…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
14.35 |
-10.66 |
0 |
3 |
0 |
43 |
386.576 |
6 |
↓
|
|
|
|
Analogs
-
31554862
-
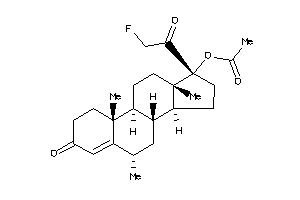
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8R,9R,10S,13S,14R,17S)-17-acetyl-10,13-dimethyl-3-oxo-2,6,7,8,9,11,12,14,15,16-decahydro-1H-cyclop
[(8R,9R,10S,13S,14R,17S)-17-acet…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
11.08 |
-13.49 |
0 |
4 |
0 |
60 |
372.505 |
3 |
↓
|
|
|
|
Analogs
-
31554862
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
11.11 |
-14.22 |
0 |
4 |
0 |
60 |
372.505 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8R,9R,10S,13S,14S,17S)-17-acetyl-10,13-dimethyl-3-oxo-2,6,7,8,9,11,12,14,15,16-decahydro-1H-cyclop
[(8R,9R,10S,13S,14S,17S)-17-acet…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
11.14 |
-14.31 |
0 |
4 |
0 |
60 |
372.505 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
11.11 |
-14.19 |
0 |
4 |
0 |
60 |
372.505 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8S,9R,10R,11S,13S,14R,17R)-11-hydroxy-17-(2-hydroxyacetyl)-10,13-dimethyl-3-oxo-2,6,7,8,9,11,12,14
[(8S,9R,10R,11S,13S,14R,17R)-11-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
5.99 |
-18.46 |
2 |
6 |
0 |
101 |
404.503 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(8S,9R,10R,11S,13S,14S,17R)-11-hydroxy-17-(2-hydroxyacetyl)-10,13-dimethyl-3-oxo-2,6,7,8,9,11,12,14
[(8S,9R,10R,11S,13S,14S,17R)-11-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
6.5 |
-19.01 |
2 |
6 |
0 |
101 |
404.503 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
4.27 |
-8.54 |
2 |
3 |
0 |
58 |
304.43 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
9.01 |
-9.8 |
2 |
3 |
0 |
58 |
416.646 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
8.8 |
-9.94 |
2 |
3 |
0 |
58 |
416.646 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8R,9S,10R,13S,14S,17R)-17-hydroxy-10,13-dimethyl-17-[2-[4-(2-pyridyl)piperazin-1-yl]acetyl]-2,6,7,8
(8R,9S,10R,13S,14S,17R)-17-hydro…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
10.5 |
-37.85 |
2 |
6 |
1 |
75 |
492.684 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.20 |
10.17 |
-12.27 |
1 |
6 |
0 |
74 |
491.676 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10S,11S,13S,14S,17S)-17-hydroxy-13-methyl-17-prop-1-ynyl-11-(4-vinylphenyl)-1,2,6,7,8,9,10,11
(8S,9R,10S,11S,13S,14S,17S)-17-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.69 |
13.46 |
-7.99 |
1 |
2 |
0 |
37 |
414.589 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10S,11S,13S,14S,17R)-17-hydroxy-13-methyl-17-prop-1-ynyl-11-(4-vinylphenyl)-1,2,6,7,8,9,10,11
(8S,9R,10S,11S,13S,14S,17R)-17-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.69 |
12.81 |
-9.84 |
1 |
2 |
0 |
37 |
414.589 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,10S,11S,13S,14R,17S)-17-hydroxy-13-methyl-17-prop-1-ynyl-11-(4-vinylphenyl)-1,2,6,7,8,9,10,11
(8S,9R,10S,11S,13S,14R,17S)-17-h…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.69 |
12.21 |
-9.33 |
1 |
2 |
0 |
37 |
414.589 |
2 |
↓
|
|