|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
739 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
9.2 |
-23.58 |
3 |
8 |
0 |
104 |
546.693 |
9 |
↓
|
|
|
|
Analogs
-
42988922
-
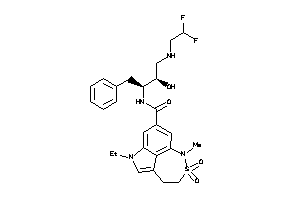
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
79 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
6.45 |
-24.14 |
3 |
8 |
0 |
104 |
552.619 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.65 |
7.86 |
-74.5 |
4 |
8 |
1 |
108 |
553.627 |
10 |
↓
|
|
|
|
Analogs
-
42988922
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
51 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
6.32 |
-24.64 |
3 |
8 |
0 |
104 |
534.629 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
7.71 |
-70.3 |
4 |
8 |
1 |
108 |
535.637 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4427 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
7.33 |
-24.03 |
3 |
8 |
0 |
104 |
602.626 |
11 |
↓
|
|
|
|
Analogs
-
42988922
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
40 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
7.53 |
-72.57 |
4 |
8 |
1 |
108 |
517.647 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.02 |
6.13 |
-23.71 |
3 |
8 |
0 |
104 |
516.639 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.31 |
Binding ≤ 10μM
|
|
BACE2-1-E |
Beta Secretase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
370 |
0.25 |
Binding ≤ 10μM
|
|
CATD-1-E |
Cathepsin D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6790 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
9.17 |
-58.12 |
4 |
8 |
1 |
108 |
511.668 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.29 |
Binding ≤ 10μM
|
|
BACE2-1-E |
Beta Secretase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1490 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
6.48 |
-23.65 |
3 |
8 |
0 |
104 |
508.644 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.89 |
7.9 |
-69.42 |
4 |
8 |
1 |
108 |
509.652 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
64 |
0.27 |
Binding ≤ 10μM
|
|
BACE2-1-E |
Beta Secretase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.24 |
Binding ≤ 10μM
|
|
CATD-1-E |
Cathepsin D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1190 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
8.71 |
-65.1 |
4 |
8 |
1 |
108 |
527.711 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BACE1-1-E |
Beta-secretase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
BACE2-1-E |
Beta Secretase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.26 |
Binding ≤ 10μM
|
|
CATD-1-E |
Cathepsin D (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
8.54 |
-76.66 |
4 |
8 |
1 |
108 |
567.654 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.92 |
7.14 |
-24.24 |
3 |
8 |
0 |
104 |
566.646 |
11 |
↓
|
|