|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
7.92 |
-54.77 |
1 |
5 |
-1 |
86 |
301.347 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.17 |
5.95 |
-117.57 |
0 |
5 |
-2 |
89 |
300.339 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.71 |
8.32 |
-61.41 |
2 |
5 |
0 |
87 |
302.355 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
8.07 |
-53.75 |
1 |
5 |
-1 |
86 |
301.347 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.40 |
6.1 |
-97.12 |
0 |
5 |
-2 |
89 |
300.339 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.94 |
8.46 |
-75.74 |
2 |
5 |
0 |
87 |
302.355 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
8.06 |
-54.1 |
1 |
5 |
-1 |
86 |
301.347 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.37 |
6.1 |
-100.48 |
0 |
5 |
-2 |
89 |
300.339 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.92 |
8.45 |
-73.52 |
2 |
5 |
0 |
87 |
302.355 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
7.31 |
-54.51 |
1 |
5 |
-1 |
86 |
287.32 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.73 |
5.34 |
-114.96 |
0 |
5 |
-2 |
89 |
286.312 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.27 |
7.74 |
-65.14 |
2 |
5 |
0 |
87 |
288.328 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
7.47 |
-53.93 |
1 |
5 |
-1 |
86 |
287.32 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.96 |
5.48 |
-95.15 |
0 |
5 |
-2 |
89 |
286.312 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.50 |
7.87 |
-79.44 |
2 |
5 |
0 |
87 |
288.328 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
7.48 |
-52.93 |
1 |
5 |
-1 |
86 |
305.31 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.05 |
5.51 |
-95.8 |
0 |
5 |
-2 |
89 |
304.302 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.59 |
7.9 |
-76.97 |
2 |
5 |
0 |
87 |
306.318 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
7.46 |
-54.22 |
1 |
5 |
-1 |
86 |
287.32 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
5.49 |
-98.46 |
0 |
5 |
-2 |
89 |
286.312 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
7.86 |
-77.15 |
2 |
5 |
0 |
87 |
288.328 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
7.38 |
-53.44 |
1 |
5 |
-1 |
86 |
305.31 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
5.4 |
-109.72 |
0 |
5 |
-2 |
89 |
304.302 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
7.81 |
-67.17 |
2 |
5 |
0 |
87 |
306.318 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
7.5 |
-56.24 |
1 |
5 |
-1 |
86 |
305.31 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
5.52 |
-100.74 |
0 |
5 |
-2 |
89 |
304.302 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
7.9 |
-78.84 |
2 |
5 |
0 |
87 |
306.318 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.10 |
6.06 |
-56.11 |
3 |
6 |
-1 |
112 |
302.335 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
6.21 |
-55.72 |
3 |
6 |
-1 |
112 |
302.335 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
6.2 |
-55.93 |
3 |
6 |
-1 |
112 |
302.335 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
4.53 |
-56.52 |
5 |
5 |
1 |
88 |
311.818 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.46 |
3.36 |
-10.26 |
4 |
5 |
0 |
84 |
310.81 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.92 |
2.58 |
-74.16 |
4 |
5 |
0 |
91 |
310.81 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
4.54 |
-56.28 |
5 |
5 |
1 |
88 |
311.818 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.46 |
3.36 |
-10.75 |
4 |
5 |
0 |
84 |
310.81 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.92 |
2.58 |
-69.38 |
4 |
5 |
0 |
91 |
310.81 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
5.38 |
-55.22 |
5 |
5 |
1 |
88 |
325.845 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.84 |
4.29 |
-10.53 |
4 |
5 |
0 |
84 |
324.837 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
3.43 |
-72.01 |
4 |
5 |
0 |
91 |
324.837 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
5.38 |
-55.02 |
5 |
5 |
1 |
88 |
325.845 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.84 |
4.27 |
-10.06 |
4 |
5 |
0 |
84 |
324.837 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
3.42 |
-67.76 |
4 |
5 |
0 |
91 |
324.837 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.21 |
2.77 |
-62.29 |
6 |
5 |
1 |
99 |
297.791 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.66 |
0.81 |
-80.3 |
5 |
5 |
0 |
103 |
296.783 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.21 |
2.76 |
-62.06 |
6 |
5 |
1 |
99 |
297.791 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.66 |
0.8 |
-74.95 |
5 |
5 |
0 |
103 |
296.783 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
4.62 |
-56.58 |
5 |
5 |
1 |
88 |
356.269 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.59 |
3.45 |
-10.11 |
4 |
5 |
0 |
84 |
355.261 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.05 |
2.66 |
-69.14 |
4 |
5 |
0 |
91 |
355.261 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
4.62 |
-56.3 |
5 |
5 |
1 |
88 |
356.269 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.59 |
3.45 |
-10.71 |
4 |
5 |
0 |
84 |
355.261 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.05 |
2.67 |
-73.83 |
4 |
5 |
0 |
91 |
355.261 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.47 |
-55.29 |
5 |
5 |
1 |
88 |
370.296 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.37 |
-10.43 |
4 |
5 |
0 |
84 |
369.288 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.42 |
3.52 |
-67.49 |
4 |
5 |
0 |
91 |
369.288 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.45 |
-55.08 |
5 |
5 |
1 |
88 |
370.296 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.97 |
4.36 |
-9.99 |
4 |
5 |
0 |
84 |
369.288 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.42 |
3.51 |
-71.75 |
4 |
5 |
0 |
91 |
369.288 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
2.86 |
-62.43 |
6 |
5 |
1 |
99 |
342.242 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.80 |
0.9 |
-74.75 |
5 |
5 |
0 |
103 |
341.234 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
2.85 |
-62.12 |
6 |
5 |
1 |
99 |
342.242 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.80 |
0.89 |
-79.97 |
5 |
5 |
0 |
103 |
341.234 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.02 |
-2.87 |
-12.05 |
1 |
6 |
0 |
61 |
380.423 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
2.21 |
-10.8 |
5 |
5 |
0 |
98 |
248.311 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.05 |
0.25 |
-50.24 |
4 |
5 |
-1 |
101 |
247.303 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
-7.64 |
-13.91 |
3 |
8 |
0 |
104 |
401.521 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
3.2 |
-18.23 |
3 |
7 |
0 |
111 |
281.293 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.47 |
13.8 |
-16.7 |
1 |
6 |
0 |
92 |
517.451 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
4.16 |
-21.54 |
2 |
7 |
0 |
101 |
325.419 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.87 |
3.16 |
-42.44 |
1 |
7 |
-1 |
107 |
324.411 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
10.42 |
-10.91 |
1 |
4 |
0 |
59 |
351.475 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.01 |
10.86 |
-36.49 |
2 |
4 |
1 |
60 |
352.483 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA-1-E |
LDL-associated Phospholipase A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4400 |
0.34 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA_HUMAN |
Q13093
|
LDL-associated Phospholipase A2, Human |
4400 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
8.27 |
-10.9 |
1 |
4 |
0 |
59 |
309.394 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.18 |
8.73 |
-36.52 |
2 |
4 |
1 |
60 |
310.402 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
11.98 |
-10.97 |
1 |
4 |
0 |
59 |
379.529 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.79 |
11.87 |
-47.71 |
2 |
4 |
1 |
60 |
380.537 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA-1-E |
LDL-associated Phospholipase A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
640 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.80 |
11.4 |
-16.06 |
1 |
6 |
0 |
89 |
456.999 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA-1-E |
LDL-associated Phospholipase A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA_HUMAN |
Q13093
|
LDL-associated Phospholipase A2, Human |
1100 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
8.25 |
-12.45 |
1 |
4 |
0 |
59 |
309.394 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.23 |
7.78 |
-43.43 |
2 |
4 |
1 |
60 |
310.402 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA-1-E |
LDL-associated Phospholipase A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1100 |
0.30 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA_HUMAN |
Q13093
|
LDL-associated Phospholipase A2, Human |
1100 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.30 |
12.76 |
-10.88 |
1 |
4 |
0 |
59 |
393.556 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.30 |
12.65 |
-48.18 |
2 |
4 |
1 |
60 |
394.564 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PAFA-1-E |
LDL-associated Phospholipase A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
780 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.27 |
16.78 |
-24.18 |
1 |
5 |
0 |
68 |
479.69 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
6.47 |
-11.69 |
1 |
4 |
0 |
59 |
281.29 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.49 |
4.37 |
-43.38 |
0 |
4 |
-1 |
62 |
280.282 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.04 |
6.93 |
-42.95 |
2 |
4 |
1 |
60 |
282.298 |
2 |
↓
|
|
|
|
Analogs
-
13982572
-
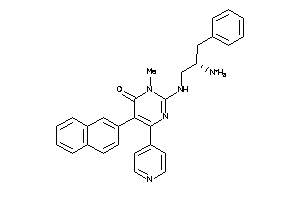
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
11.34 |
-11.26 |
1 |
5 |
0 |
60 |
352.413 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.63 |
11.8 |
-42.67 |
2 |
5 |
1 |
61 |
353.421 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
12.8 |
-11.11 |
1 |
5 |
0 |
60 |
386.43 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.59 |
13.26 |
-43.43 |
2 |
5 |
1 |
61 |
387.438 |
5 |
↓
|
|
|
|
Analogs
-
13982572
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
12.37 |
-64.11 |
4 |
6 |
1 |
87 |
444.534 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.84 |
12 |
-11.28 |
3 |
6 |
0 |
86 |
443.526 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.84 |
12.83 |
-112.83 |
5 |
6 |
2 |
89 |
445.542 |
7 |
↓
|
|
|
|
Analogs
-
13982572
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
12.4 |
-62.75 |
4 |
6 |
1 |
87 |
444.534 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.84 |
12.17 |
-12.78 |
3 |
6 |
0 |
86 |
443.526 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.84 |
12.86 |
-111.24 |
5 |
6 |
2 |
89 |
445.542 |
7 |
↓
|
|
|
|
Analogs
-
13982572
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
12.21 |
-65.77 |
4 |
6 |
1 |
87 |
444.534 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.05 |
11.94 |
-12.09 |
3 |
6 |
0 |
86 |
443.526 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.05 |
12.66 |
-119.46 |
5 |
6 |
2 |
89 |
445.542 |
7 |
↓
|
|
|
|
Analogs
-
13982572
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
12.2 |
-63.99 |
4 |
6 |
1 |
87 |
444.534 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.05 |
11.89 |
-12.74 |
3 |
6 |
0 |
86 |
443.526 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.05 |
12.66 |
-117.08 |
5 |
6 |
2 |
89 |
445.542 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
4.78 |
-11.52 |
2 |
5 |
0 |
82 |
300.387 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.60 |
5.72 |
-55 |
3 |
5 |
1 |
86 |
301.395 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
4.6 |
-11.93 |
2 |
5 |
0 |
82 |
300.387 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.60 |
5.72 |
-55.22 |
3 |
5 |
1 |
86 |
301.395 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
1.92 |
-13.82 |
3 |
5 |
0 |
96 |
238.316 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.27 |
2.18 |
-64.44 |
4 |
5 |
1 |
97 |
239.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
1.91 |
-12.49 |
3 |
5 |
0 |
96 |
238.316 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.27 |
2.14 |
-64.54 |
4 |
5 |
1 |
97 |
239.324 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
2.19 |
-12.52 |
2 |
5 |
0 |
82 |
252.343 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.11 |
1.5 |
-78.8 |
2 |
5 |
0 |
89 |
252.343 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.65 |
3.48 |
-60.3 |
3 |
5 |
1 |
86 |
253.351 |
5 |
↓
|
|