|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4480 |
0.42 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT_HUMAN |
Q16769
|
Glutaminyl-peptide Cyclotransferase, Human |
4480 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
8.51 |
-15.93 |
2 |
4 |
0 |
42 |
260.366 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2030 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT_HUMAN |
Q16769
|
Glutaminyl-peptide Cyclotransferase, Human |
2030 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
9.44 |
-44.56 |
3 |
5 |
1 |
46 |
304.443 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.40 |
8.92 |
-16.78 |
2 |
5 |
0 |
45 |
303.435 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.40 |
10.2 |
-87.18 |
4 |
5 |
0 |
48 |
305.451 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1660 |
0.40 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT_HUMAN |
Q16769
|
Glutaminyl-peptide Cyclotransferase, Human |
1660 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
10.05 |
-45.05 |
3 |
4 |
1 |
43 |
307.468 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.74 |
9.54 |
-16.43 |
2 |
4 |
0 |
42 |
306.46 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1860 |
0.40 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT_HUMAN |
Q16769
|
Glutaminyl-peptide Cyclotransferase, Human |
1860 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
8.29 |
-44.24 |
3 |
5 |
1 |
52 |
291.4 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.33 |
7.77 |
-16.24 |
2 |
5 |
0 |
51 |
290.392 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
750 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
7.6 |
-46.58 |
3 |
6 |
1 |
62 |
321.426 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
7.08 |
-18.65 |
2 |
6 |
0 |
60 |
320.418 |
9 |
↓
|
|
|
|
Analogs
-
39973988
-
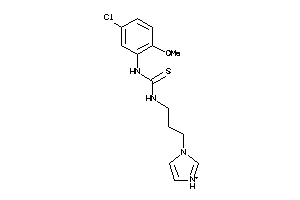
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
560 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
7.72 |
-44.24 |
3 |
6 |
1 |
62 |
321.426 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.34 |
7.2 |
-16.28 |
2 |
6 |
0 |
60 |
320.418 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
8.15 |
-49.98 |
3 |
7 |
1 |
71 |
351.452 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.93 |
7.63 |
-20.44 |
2 |
7 |
0 |
70 |
350.444 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.95 |
8.23 |
-47.89 |
3 |
6 |
1 |
62 |
321.426 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.95 |
7.72 |
-18.89 |
2 |
6 |
0 |
60 |
320.418 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
8.47 |
-46.72 |
3 |
6 |
1 |
62 |
335.453 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.42 |
7.96 |
-18 |
2 |
6 |
0 |
60 |
334.445 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
760 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
8.76 |
-47.25 |
3 |
6 |
1 |
62 |
335.453 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.42 |
8.25 |
-18.08 |
2 |
6 |
0 |
60 |
334.445 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4830 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT_HUMAN |
Q16769
|
Glutaminyl-peptide Cyclotransferase, Human |
4830 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
9.73 |
-20.11 |
1 |
6 |
0 |
52 |
334.445 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT-1-E |
Glutaminyl-peptide Cyclotransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1790 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
QPCT_HUMAN |
Q16769
|
Glutaminyl-peptide Cyclotransferase, Human |
1790 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
9.5 |
-48.18 |
3 |
5 |
1 |
60 |
303.411 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
8.99 |
-18.85 |
2 |
5 |
0 |
59 |
302.403 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
-1.21 |
-15.38 |
2 |
4 |
0 |
41 |
288.42 |
8 |
↓
|
|