|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Binding ≤ 10μM
|
|
TOP3-1-B |
DNA Topoisomerase III (cluster #1 Of 1), Bacterial |
Bacteria |
420 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.86 |
4.04 |
-9.12 |
3 |
5 |
0 |
78 |
243.266 |
2 |
↓
|
|
|
|
Analogs
-
26908
-
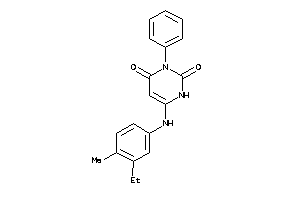
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1340 |
0.43 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1340 |
0.43 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1340 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
6.55 |
-9.41 |
2 |
5 |
0 |
67 |
257.293 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.46 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.46 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
7.54 |
-8.99 |
2 |
5 |
0 |
67 |
271.32 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
8.1 |
-9.24 |
2 |
5 |
0 |
67 |
283.331 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.43 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.43 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
8.3 |
-9.05 |
2 |
5 |
0 |
67 |
285.347 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.45 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.45 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
9.09 |
-9.02 |
2 |
5 |
0 |
67 |
299.374 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RPC10-1-E |
DNA-directed RNA Polymerases III 12.5 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
240 |
0.39 |
Binding ≤ 10μM
|
|
RPC4-1-E |
DNA-directed RNA Polymerase III 47 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
240 |
0.39 |
Binding ≤ 10μM
|
|
RPC6-1-E |
DNA-directed RNA Polymerase III 39 KDa Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
240 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
10.65 |
-8.9 |
2 |
5 |
0 |
67 |
327.428 |
7 |
↓
|
|