|
|
Analogs
-
42891931
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
0.22 |
-14.11 |
1 |
6 |
0 |
67 |
423.605 |
3 |
↓
|
|
|
|
Analogs
-
42891931
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
0.38 |
-13.92 |
1 |
6 |
0 |
67 |
423.605 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
8.3 |
-5.15 |
1 |
2 |
0 |
29 |
249.398 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
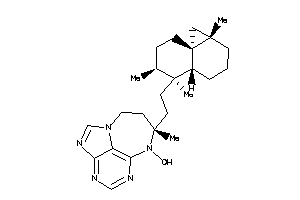
Popular Name:
(2E)-2-[2-[(1S,2S,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]ethylidene]butanedial
(2E)-2-[2-[(1S,2S,4aS,8aR)-1,2,4…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
1600 |
0.37 |
Functional ≤ 10μM |
|
Z80362-4-O |
P388 (Lymphoma Cells) (cluster #4 Of 8), Other |
Other |
3300 |
0.35 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
3300 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.19 |
9.49 |
-11.43 |
0 |
2 |
0 |
34 |
302.458 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-5-[(1S,2R,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-3-(acetoxymethyl)-4-hydroxy-pent-
[(Z)-5-[(1S,2R,4aS,8aR)-1,2,4a-t…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
2500 |
0.27 |
Functional ≤ 10μM |
|
Z80362-4-O |
P388 (Lymphoma Cells) (cluster #4 Of 8), Other |
Other |
2500 |
0.27 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
2500 |
0.27 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
13.24 |
-12.27 |
0 |
5 |
0 |
70 |
406.563 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(Z)-5-[(1S,2S,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-3-(acetoxymethyl)-4-hydroxy-pent-
[(Z)-5-[(1S,2S,4aS,8aR)-1,2,4a-t…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
2500 |
0.27 |
Functional ≤ 10μM |
|
Z80362-4-O |
P388 (Lymphoma Cells) (cluster #4 Of 8), Other |
Other |
2500 |
0.27 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
2500 |
0.27 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
12.82 |
-12.21 |
0 |
5 |
0 |
70 |
406.563 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E)-5-[(1S,2R,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-3-acetyl-pent-3-enal
(E)-5-[(1S,2R,4aS,8aR)-1,2,4a-tr…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM |
|
Z80362-4-O |
P388 (Lymphoma Cells) (cluster #4 Of 8), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
10.78 |
-7.66 |
0 |
2 |
0 |
34 |
316.485 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(E)-5-[(1S,2S,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-3-acetyl-pent-3-enal
(E)-5-[(1S,2S,4aS,8aR)-1,2,4a-tr…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM |
|
Z80362-4-O |
P388 (Lymphoma Cells) (cluster #4 Of 8), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM |
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
1600 |
0.35 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
10.25 |
-8 |
0 |
2 |
0 |
34 |
316.485 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3E)-3-[2-[(1S,2R,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]ethylidene]butane-1,2,4-trio
(2S,3E)-3-[2-[(1S,2R,4aS,8aR)-1,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
3700 |
0.33 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
3700 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
2.43 |
-6.61 |
3 |
3 |
0 |
61 |
322.489 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3E)-3-[2-[(1S,2S,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]ethylidene]butane-1,2,4-trio
(2S,3E)-3-[2-[(1S,2S,4aS,8aR)-1,…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166-10-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #10 Of 12), Other |
Other |
3700 |
0.33 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
3700 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
1.9 |
-5.62 |
3 |
3 |
0 |
61 |
322.489 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E)-4-[(1S,2R,4aR,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-2-[(1S)-1,2-diacetoxyethyl]but-2-
[(E)-4-[(1S,2R,4aR,8aR)-1,2,4a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
15.3 |
-12.83 |
0 |
6 |
0 |
79 |
448.6 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E)-4-[(1S,2S,4aR,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-2-[(1S)-1,2-diacetoxyethyl]but-2-
[(E)-4-[(1S,2S,4aR,8aR)-1,2,4a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
15.23 |
-12.48 |
0 |
6 |
0 |
79 |
448.6 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E)-4-[(1S,2R,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-2-[(1S)-1,2-diacetoxyethyl]but-2-
[(E)-4-[(1S,2R,4aS,8aR)-1,2,4a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
15.35 |
-13.18 |
0 |
6 |
0 |
79 |
448.6 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(E)-4-[(1S,2S,4aS,8aR)-1,2,4a-trimethyl-5-methylene-decalin-1-yl]-2-[(1S)-1,2-diacetoxyethyl]but-2-
[(E)-4-[(1S,2S,4aS,8aR)-1,2,4a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.48 |
14.43 |
-12.13 |
0 |
6 |
0 |
79 |
448.6 |
11 |
↓
|
|
|
|
Analogs
-
28639179
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
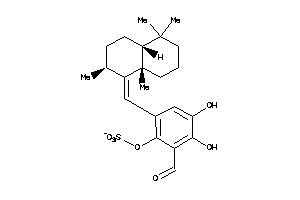
[6-[(Z)-[(2S,4aS,8aS)-2,5,5,8a-tetramethyldecalin-1-ylidene]methyl]-2-formyl-3,4-dihydroxy-phenyl]
[6-[(Z)-[(2S,4aS,8aS)-2,5,5,8a-t…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
4.63 |
-49.34 |
2 |
7 |
-1 |
124 |
437.534 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.64 |
5.69 |
-115.75 |
1 |
7 |
-2 |
127 |
436.526 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(2R,4aS,5R,6S,8aR)-5-[(E)-5-(6-amino-9-methyl-purin-9-ium-7-yl)-3-methyl-pent-3-enyl]-5,6,8a-trimeth
(2R,4aS,5R,6S,8aR)-5-[(E)-5-(6-a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
13.31 |
-38.12 |
3 |
6 |
1 |
81 |
438.64 |
5 |
↓
|
|