|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR1-1-E |
C-C Chemokine Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
316 |
0.34 |
Binding ≤ 10μM |
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
251 |
0.34 |
Binding ≤ 10μM |
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1995 |
0.30 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.03 |
9.04 |
-59.95 |
1 |
6 |
-1 |
96 |
402.835 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.03 |
9.12 |
-119.1 |
0 |
6 |
-2 |
98 |
401.827 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3162 |
0.28 |
Binding ≤ 10μM |
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3162 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
9.69 |
-60.28 |
1 |
6 |
-1 |
96 |
416.862 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.43 |
9.73 |
-119.82 |
0 |
6 |
-2 |
98 |
415.854 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCR2-1-E |
C-C Chemokine Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.41 |
Binding ≤ 10μM |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7943 |
0.25 |
Binding ≤ 10μM |
|
CCR4-1-E |
C-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
398 |
0.31 |
Functional ≤ 10μM |
|
CCR5-1-E |
C-C Chemokine Receptor Type 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
631 |
0.30 |
Functional ≤ 10μM |
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4169 |
0.26 |
ADME/T ≤ 10μM |
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.24 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.29 |
10.1 |
-112.42 |
0 |
6 |
-2 |
98 |
470.717 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.29 |
10.01 |
-56.2 |
1 |
6 |
-1 |
96 |
471.725 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
10.13 |
-110.47 |
0 |
7 |
-2 |
107 |
471.918 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.78 |
7.72 |
-108.26 |
0 |
7 |
-2 |
107 |
449.843 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
8.75 |
-108.85 |
0 |
8 |
-2 |
116 |
487.917 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
8.64 |
-108 |
0 |
7 |
-2 |
107 |
463.87 |
8 |
↓
|
|
|
|
Analogs
-
13059297
-
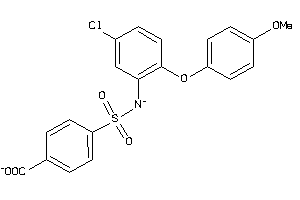
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
7.44 |
-49.92 |
1 |
6 |
-1 |
96 |
402.835 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
7.34 |
-59.42 |
1 |
8 |
-1 |
114 |
442.469 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.24 |
7.44 |
-132.65 |
0 |
8 |
-2 |
116 |
441.461 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
7.2 |
-53.95 |
1 |
7 |
-1 |
105 |
398.416 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.30 |
7.71 |
-141.53 |
0 |
7 |
-2 |
107 |
397.408 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.52 |
-61.76 |
1 |
6 |
-1 |
96 |
386.38 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.41 |
7.94 |
-137.95 |
0 |
6 |
-2 |
98 |
385.372 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
8.28 |
-123.56 |
0 |
7 |
-2 |
121 |
392.392 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.05 |
8.23 |
-53.69 |
1 |
7 |
-1 |
119 |
393.4 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
7.8 |
-15.75 |
2 |
7 |
0 |
94 |
440.521 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.81 |
7.87 |
-45.83 |
1 |
7 |
-1 |
96 |
439.513 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
7.5 |
-14.41 |
2 |
7 |
0 |
94 |
460.939 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
8.31 |
-14.36 |
2 |
7 |
0 |
94 |
474.966 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
6.66 |
-14.84 |
2 |
7 |
0 |
94 |
446.912 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
7.76 |
-12.29 |
1 |
7 |
0 |
91 |
447.896 |
8 |
↓
|
|
|
|
Analogs
-
5134981
-
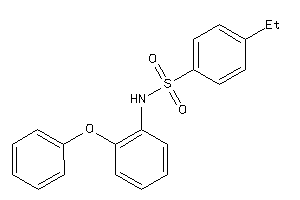
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
9.53 |
-9.67 |
1 |
4 |
0 |
55 |
367.47 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.71 |
9.6 |
-46.18 |
0 |
4 |
-1 |
57 |
366.462 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.27 |
10.27 |
-9.66 |
1 |
4 |
0 |
55 |
381.497 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.27 |
10.34 |
-49.43 |
0 |
4 |
-1 |
57 |
380.489 |
8 |
↓
|
|
|
|
Analogs
-
5383900
-
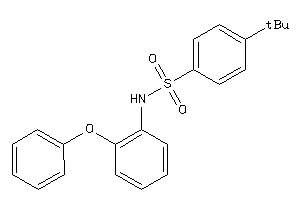
-
3435884
-
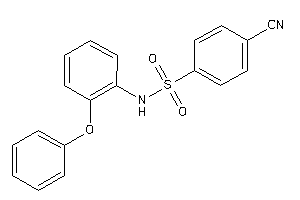
-
6328360
-
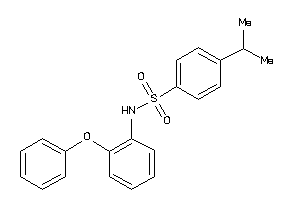
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
9.96 |
-9.6 |
1 |
4 |
0 |
55 |
381.497 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.71 |
10.03 |
-49.58 |
0 |
4 |
-1 |
57 |
380.489 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.23 |
8.79 |
-9.81 |
1 |
4 |
0 |
55 |
353.443 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.23 |
8.87 |
-45.94 |
0 |
4 |
-1 |
57 |
352.435 |
5 |
↓
|
|
|
|
Analogs
-
14257158
-
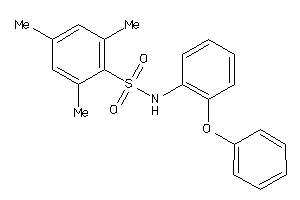
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.23 |
8.82 |
-9.73 |
1 |
4 |
0 |
55 |
353.443 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.23 |
8.88 |
-46 |
0 |
4 |
-1 |
57 |
352.435 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
7.05 |
-9.07 |
1 |
4 |
0 |
55 |
361.369 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.67 |
7.13 |
-42.76 |
0 |
4 |
-1 |
57 |
360.361 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
7.04 |
-10.07 |
1 |
4 |
0 |
55 |
361.369 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.67 |
7.14 |
-45.8 |
0 |
4 |
-1 |
57 |
360.361 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.36 |
10.42 |
-9.84 |
1 |
4 |
0 |
55 |
395.524 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.36 |
10.51 |
-50.26 |
0 |
4 |
-1 |
57 |
394.516 |
5 |
↓
|
|
|
|
Analogs
-
14257150
-
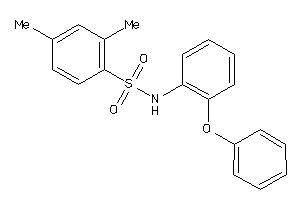
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.61 |
9.57 |
-9.08 |
1 |
4 |
0 |
55 |
367.47 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.61 |
9.66 |
-47.56 |
0 |
4 |
-1 |
57 |
366.462 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.30 |
8.43 |
-8.38 |
1 |
4 |
0 |
55 |
428.724 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.30 |
8.53 |
-39.62 |
0 |
4 |
-1 |
57 |
427.716 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
9.23 |
-8.46 |
1 |
4 |
0 |
55 |
387.888 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.84 |
9.31 |
-41.43 |
0 |
4 |
-1 |
57 |
386.88 |
5 |
↓
|
|