|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.57 |
-1.29 |
-8.16 |
1 |
3 |
0 |
38 |
212.252 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.57 |
-1.24 |
-30.66 |
2 |
3 |
1 |
39 |
213.26 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z103196-1-O |
22RV1 (cluster #1 Of 2), Other |
Other |
8800 |
0.26 |
Functional ≤ 10μM |
|
Z103200-1-O |
769-P (cluster #1 Of 1), Other |
Other |
2200 |
0.29 |
Functional ≤ 10μM |
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
2600 |
0.29 |
Functional ≤ 10μM |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
4600 |
0.28 |
Functional ≤ 10μM |
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 6), Other |
Other |
6200 |
0.27 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.95 |
10.07 |
-31.4 |
2 |
4 |
1 |
34 |
367.561 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.95 |
11.78 |
-35.77 |
2 |
4 |
1 |
34 |
367.561 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.95 |
12.19 |
-79.89 |
3 |
4 |
2 |
36 |
368.569 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
7100 |
0.18 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547_9HIV1 |
Q72547
|
Human Immunodeficiency Virus Type 1 Reverse Transcriptase, 9hiv1 |
7100 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
13.45 |
-19.38 |
2 |
9 |
0 |
131 |
524.577 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.05 |
13.81 |
-38.2 |
3 |
9 |
1 |
132 |
525.585 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.73 |
13.6 |
-11.45 |
1 |
4 |
0 |
51 |
450.925 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.73 |
13.85 |
-32.51 |
2 |
4 |
1 |
52 |
451.933 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.67 |
15.12 |
-8.75 |
1 |
3 |
0 |
42 |
489.789 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.67 |
15.38 |
-33.1 |
2 |
3 |
1 |
43 |
490.797 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
6.7 |
-9.18 |
1 |
3 |
0 |
42 |
234.258 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.19 |
6.97 |
-31.49 |
2 |
3 |
1 |
43 |
235.266 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
7.89 |
-8.02 |
2 |
3 |
0 |
44 |
259.312 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
6.79 |
-10.06 |
2 |
4 |
0 |
50 |
275.311 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.29 |
13.99 |
-23.57 |
0 |
4 |
0 |
36 |
398.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
7.85 |
-14.63 |
2 |
7 |
0 |
87 |
426.476 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.17 |
7.9 |
-37.95 |
3 |
7 |
1 |
89 |
427.484 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
10.3 |
-28.44 |
1 |
4 |
1 |
45 |
297.378 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.39 |
9.9 |
-9.74 |
0 |
4 |
0 |
44 |
296.37 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
7.95 |
-29.06 |
1 |
4 |
1 |
45 |
255.297 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.95 |
7.56 |
-10.66 |
0 |
4 |
0 |
44 |
254.289 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
11.09 |
-10.86 |
0 |
4 |
0 |
44 |
330.387 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.54 |
11.48 |
-30.96 |
1 |
4 |
1 |
45 |
331.395 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
11.53 |
-10.21 |
0 |
4 |
0 |
38 |
367.518 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.38 |
11.87 |
-32.73 |
1 |
4 |
1 |
39 |
368.526 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
12.05 |
-10.28 |
0 |
4 |
0 |
38 |
379.529 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.62 |
12.21 |
-33.22 |
1 |
4 |
1 |
39 |
380.537 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
8.03 |
-10.12 |
1 |
4 |
0 |
49 |
311.41 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.88 |
8.14 |
-33.46 |
2 |
4 |
1 |
50 |
312.418 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
12.95 |
-11.64 |
0 |
4 |
0 |
38 |
401.535 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.54 |
13.1 |
-36.47 |
1 |
4 |
1 |
39 |
402.543 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
8.27 |
-10.78 |
1 |
4 |
0 |
49 |
293.37 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.40 |
8.33 |
-32.43 |
2 |
4 |
1 |
50 |
294.378 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
10.8 |
-11.12 |
0 |
4 |
0 |
38 |
349.459 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.10 |
10.96 |
-33.72 |
1 |
4 |
1 |
39 |
350.467 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.70 |
13.02 |
-13.42 |
0 |
4 |
0 |
38 |
419.525 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.70 |
13.17 |
-40.56 |
1 |
4 |
1 |
39 |
420.533 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.11 |
11.39 |
-17 |
4 |
8 |
0 |
116 |
580.729 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.11 |
11.46 |
-33.52 |
5 |
8 |
1 |
118 |
581.737 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.20 |
12.83 |
-25.34 |
2 |
9 |
0 |
119 |
628.795 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.20 |
12.9 |
-44.31 |
3 |
9 |
1 |
121 |
629.803 |
10 |
↓
|
|
|
|
Analogs
-
35454571
-
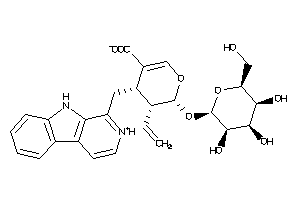
-
35454575
-
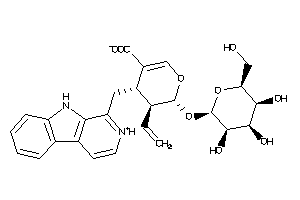
-
35454579
-
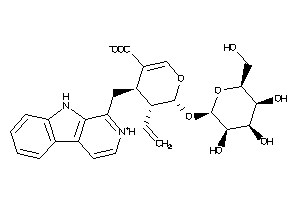
-
35454583
-
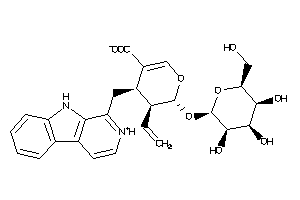
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S)-4-(9H-pyrido[3,4-b]indol-1-ylmethyl)-6-[(2S,3S,4S,5S,6S)-3,4,5-trihydroxy-6-(hydroxymethy
(4S,5S,6S)-4-(9H-pyrido[3,4-b]in…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
0.14 |
-62.95 |
5 |
11 |
-1 |
177 |
511.507 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
0.38 |
-33.22 |
6 |
11 |
0 |
179 |
512.515 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-0.67 |
-46.71 |
7 |
11 |
1 |
176 |
513.523 |
7 |
↓
|
|
|
|
Analogs
-
35454571
-

-
35454575
-

-
35454579
-

-
35454583
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S)-4-(9H-pyrido[3,4-b]indol-1-ylmethyl)-6-[(2S,3S,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethy
(4S,5S,6S)-4-(9H-pyrido[3,4-b]in…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
-0.69 |
-56.35 |
5 |
11 |
-1 |
177 |
511.507 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-0.45 |
-42.32 |
6 |
11 |
0 |
179 |
512.515 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-1.47 |
-49.65 |
7 |
11 |
1 |
176 |
513.523 |
7 |
↓
|
|
|
|
Analogs
-
35454571
-

-
35454575
-

-
35454579
-

-
35454583
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S)-4-(9H-pyrido[3,4-b]indol-1-ylmethyl)-6-[(2R,3S,4S,5S,6S)-3,4,5-trihydroxy-6-(hydroxymethy
(4S,5S,6S)-4-(9H-pyrido[3,4-b]in…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
-0.66 |
-61.26 |
5 |
11 |
-1 |
177 |
511.507 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-0.43 |
-47.95 |
6 |
11 |
0 |
179 |
512.515 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-1.55 |
-52.93 |
7 |
11 |
1 |
176 |
513.523 |
7 |
↓
|
|
|
|
Analogs
-
35454571
-

-
35454575
-

-
35454579
-

-
35454583
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S)-4-(9H-pyrido[3,4-b]indol-1-ylmethyl)-6-[(2R,3S,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethy
(4S,5S,6S)-4-(9H-pyrido[3,4-b]in…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
-0.69 |
-60.47 |
5 |
11 |
-1 |
177 |
511.507 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-0.46 |
-48.52 |
6 |
11 |
0 |
179 |
512.515 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.02 |
-1.39 |
-51.68 |
7 |
11 |
1 |
176 |
513.523 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.25 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
124 |
0.23 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.23 |
Binding ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
30 |
0.25 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
30 |
0.25 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
30 |
0.25 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
124 |
0.23 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
100 |
0.23 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
100 |
0.23 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
100 |
0.23 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
30 |
0.25 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
124 |
0.23 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
100 |
0.23 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
100 |
0.23 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
100 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.14 |
15.89 |
-17.76 |
2 |
8 |
0 |
110 |
558.638 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.14 |
16.34 |
-44.31 |
3 |
8 |
1 |
111 |
559.646 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.14 |
16.79 |
-76.81 |
4 |
8 |
2 |
112 |
560.654 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
9.86 |
-9.79 |
1 |
3 |
0 |
46 |
351.203 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.85 |
9.28 |
-35.94 |
2 |
3 |
1 |
47 |
352.211 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
6.23 |
-36.14 |
3 |
4 |
1 |
59 |
305.357 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
5.55 |
-9.26 |
2 |
4 |
0 |
58 |
304.349 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
6.27 |
-35.94 |
3 |
4 |
1 |
59 |
305.357 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.59 |
5.59 |
-9.26 |
2 |
4 |
0 |
58 |
304.349 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
10.49 |
-48.61 |
1 |
8 |
-1 |
132 |
360.305 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.05 |
9.89 |
-30.15 |
2 |
8 |
0 |
133 |
361.313 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
9.69 |
-60.9 |
1 |
8 |
-1 |
132 |
360.305 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.01 |
9.98 |
-29.99 |
2 |
8 |
0 |
133 |
361.313 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
9.92 |
-10.05 |
1 |
3 |
0 |
46 |
286.334 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.51 |
9.33 |
-32.93 |
2 |
3 |
1 |
47 |
287.342 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
9.91 |
-10.24 |
1 |
3 |
0 |
46 |
286.334 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.49 |
9.34 |
-32.88 |
2 |
3 |
1 |
47 |
287.342 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
9.87 |
-8.32 |
1 |
3 |
0 |
46 |
351.203 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.88 |
9.29 |
-34.88 |
2 |
3 |
1 |
47 |
352.211 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.83 |
9.86 |
-10.74 |
1 |
3 |
0 |
46 |
351.203 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.83 |
9.57 |
-13.81 |
2 |
3 |
0 |
47 |
352.211 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
8.57 |
-10.23 |
1 |
4 |
0 |
55 |
302.333 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.12 |
7.99 |
-34.17 |
2 |
4 |
1 |
56 |
303.341 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
9.96 |
-9.9 |
1 |
6 |
0 |
92 |
317.304 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.03 |
9.38 |
-41.75 |
2 |
6 |
1 |
93 |
318.312 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
9.95 |
-13.74 |
1 |
6 |
0 |
92 |
317.304 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.00 |
9.37 |
-44.24 |
2 |
6 |
1 |
93 |
318.312 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
7.61 |
-33.8 |
3 |
3 |
1 |
50 |
289.358 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.98 |
6.93 |
-8.07 |
2 |
3 |
0 |
49 |
288.35 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.98 |
7.64 |
-33.68 |
3 |
3 |
1 |
50 |
289.358 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.98 |
6.97 |
-7.99 |
2 |
3 |
0 |
49 |
288.35 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
7.6 |
-36.8 |
3 |
3 |
1 |
50 |
354.227 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.34 |
6.9 |
-7.65 |
2 |
3 |
0 |
49 |
353.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
7.56 |
-36.94 |
3 |
3 |
1 |
50 |
354.227 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.34 |
6.88 |
-7.66 |
2 |
3 |
0 |
49 |
353.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
9.78 |
-51.47 |
1 |
5 |
-1 |
86 |
315.308 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.09 |
9.17 |
-26.5 |
2 |
5 |
0 |
87 |
316.316 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
7.33 |
-29.56 |
3 |
3 |
1 |
42 |
240.33 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.69 |
6.88 |
-6.3 |
2 |
3 |
0 |
41 |
239.322 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.27 |
8.02 |
-29.62 |
3 |
3 |
1 |
42 |
254.357 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.27 |
7.56 |
-6.09 |
2 |
3 |
0 |
41 |
253.349 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.86 |
9.24 |
-30.57 |
3 |
3 |
1 |
42 |
280.395 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.86 |
8.79 |
-6.12 |
2 |
3 |
0 |
41 |
279.387 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
9.63 |
-32.5 |
3 |
3 |
1 |
42 |
288.374 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.38 |
9.18 |
-7.5 |
2 |
3 |
0 |
41 |
287.366 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
8.55 |
-10.59 |
1 |
4 |
0 |
55 |
302.333 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.10 |
7.96 |
-32.84 |
2 |
4 |
1 |
56 |
303.341 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
10.6 |
-10.41 |
0 |
3 |
0 |
35 |
286.334 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.36 |
11.07 |
-36.67 |
1 |
3 |
1 |
36 |
287.342 |
1 |
↓
|
|