|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
7.12 |
-49.97 |
1 |
4 |
-1 |
73 |
260.313 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
0.19 |
-18.2 |
5 |
8 |
0 |
134 |
338.408 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
-0.04 |
-18.33 |
5 |
8 |
0 |
134 |
338.408 |
4 |
↓
|
|
|
|
Analogs
-
12410098
-
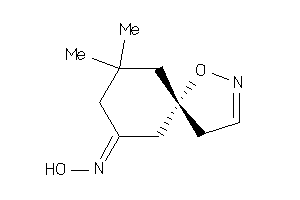
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
2.45 |
-11.23 |
1 |
4 |
0 |
54 |
196.25 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
1.78 |
2.44 |
-11.63 |
1 |
4 |
0 |
54 |
196.25 |
0 |
↓
|
|
|
|
Analogs
-
11536009
-
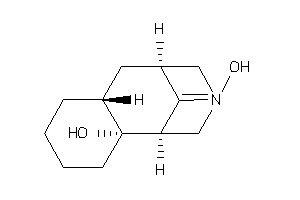
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
1.78 |
-7.1 |
2 |
3 |
0 |
53 |
223.316 |
0 |
↓
|
|
|
|
Analogs
-
11536008
-
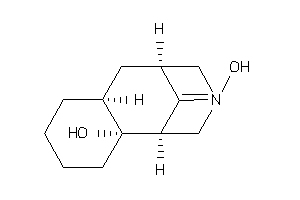
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
2.25 |
-6.81 |
2 |
3 |
0 |
53 |
223.316 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
3.87 |
-16.22 |
2 |
6 |
0 |
88 |
323.418 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
4.1 |
-54.89 |
1 |
6 |
-1 |
90 |
322.41 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
4.09 |
-56.79 |
1 |
6 |
-1 |
90 |
322.41 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3aS,5aS,6E,8R,9S,9aS,9bR)-2-tert-butyl-6-[(2S)-3-(2-furylmethoxy)-2-hydroxy-propoxy]imino-8,9-dihyd
(3aS,5aS,6E,8R,9S,9aS,9bR)-2-ter…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
2.12 |
-18.85 |
3 |
10 |
0 |
142 |
478.542 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.26 |
15.55 |
-19.94 |
0 |
8 |
0 |
92 |
473.577 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.26 |
15.59 |
-18.32 |
0 |
8 |
0 |
92 |
473.577 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3E,5S,6S,8S,9R,10R,13S,14S,17S)-3-(2-aminoethoxyimino)-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,1
(3E,5S,6S,8S,9R,10R,13S,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
2.26 |
-43.42 |
5 |
5 |
1 |
90 |
365.538 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3E,5S,6S,8S,9S,10R,13S,14S,17S)-3-(2-aminoethoxyimino)-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,1
(3E,5S,6S,8S,9S,10R,13S,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
2.17 |
-43.45 |
5 |
5 |
1 |
90 |
365.538 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3E,5S,6S,8R,9R,10R,13S,14S,17S)-3-(2-aminoethoxyimino)-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,1
(3E,5S,6S,8R,9R,10R,13S,14S,17S)…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT1A1-2-E |
Sodium/potassium-transporting ATPase Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A2-2-E |
Sodium/potassium-transporting ATPase Alpha-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A3-2-E |
Sodium/potassium-transporting ATPase Alpha-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1A4-2-E |
Sodium/potassium-transporting ATPase Alpha-4 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B1-2-E |
Sodium/potassium-transporting ATPase Beta-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B2-2-E |
Sodium/potassium-transporting ATPase Beta-2 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
AT1B3-2-E |
Sodium/potassium-transporting ATPase Beta-3 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
|
ATNG-1-E |
Sodium/potassium-transporting ATPase Gamma Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
2.34 |
-43.1 |
5 |
5 |
1 |
90 |
365.538 |
3 |
↓
|
|
|
|
Analogs
-
19865172
-
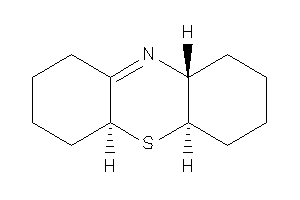
-
36372548
-
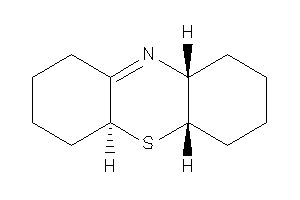
-
36372549
-
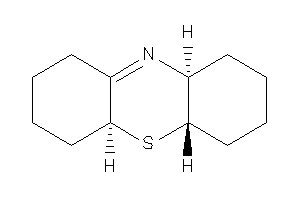
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,5aR,9aS)-2,3,4,4a,5a,6,7,8,9,9a-decahydro-1H-phenothiazine
(4aS,5aR,9aS)-2,3,4,4a,5a,6,7,8,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
6.48 |
-2.8 |
0 |
1 |
0 |
12 |
209.358 |
0 |
↓
|
|
|
|
Analogs
-
36372548
-

-
36372549
-

-
19865169
-
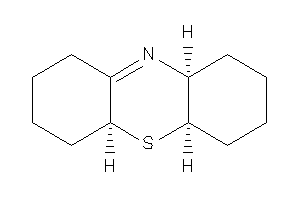
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aS,5aR,9aR)-2,3,4,4a,5a,6,7,8,9,9a-decahydro-1H-phenothiazine
(4aS,5aR,9aR)-2,3,4,4a,5a,6,7,8,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
6.59 |
-3.21 |
0 |
1 |
0 |
12 |
209.358 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
4-fluoro-N-[1,4,5,6,7,8-hexahydrospiro(2H-3,1-benzothiazine-4,1'-cyclohexane)-2-ylidene]benzamide
4-fluoro-N-[1,4,5,6,7,8-hexahydr…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.50 |
9.1 |
-37.35 |
1 |
3 |
1 |
43 |
359.49 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.71 |
9.05 |
-8.22 |
1 |
3 |
0 |
41 |
358.482 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.55 |
10.03 |
-8.51 |
1 |
3 |
0 |
41 |
358.482 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
18.6 |
-23.52 |
1 |
3 |
1 |
28 |
452.647 |
5 |
↓
|
|
|
|
Analogs
-
36143485
-
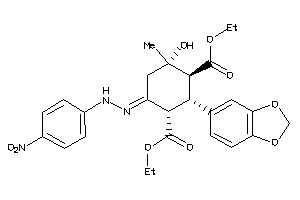
-
33922129
-
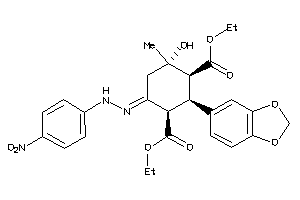
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.31 |
10.68 |
-24.94 |
2 |
12 |
0 |
162 |
527.53 |
10 |
↓
|
|
|
|
Analogs
-
33922129
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.31 |
10.37 |
-17.43 |
2 |
12 |
0 |
162 |
527.53 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
7.88 |
-16.66 |
3 |
9 |
0 |
114 |
402.48 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
8.27 |
-17.7 |
3 |
9 |
0 |
114 |
402.48 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
39702236
-
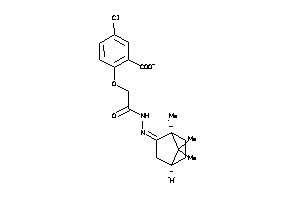
-
6269351
-
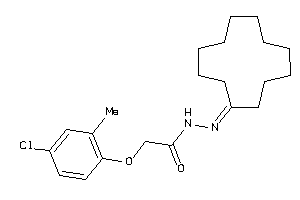
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.87 |
-15.67 |
1 |
4 |
0 |
51 |
308.809 |
4 |
↓
|
|
|
|
Analogs
-
39702236
-

-
6269351
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.87 |
-15.84 |
1 |
4 |
0 |
51 |
308.809 |
4 |
↓
|
|
|
|
Analogs
-
12402552
-
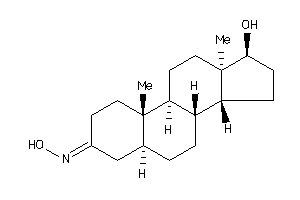
-
12402553
-
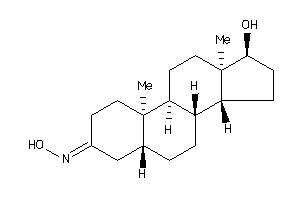
-
12402554
-
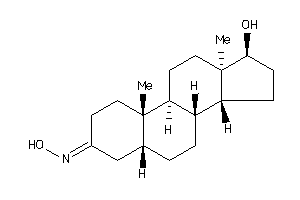
-
12877346
-
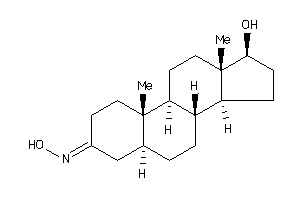
-
13146629
-
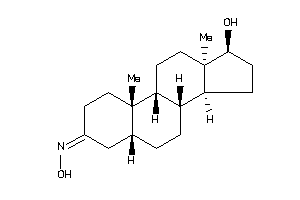
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,8R,9S,10R,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5S,8R,9S,10R,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.74 |
-6.24 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
12402553
-

-
12402554
-

-
12877346
-

-
13146629
-

-
13146631
-
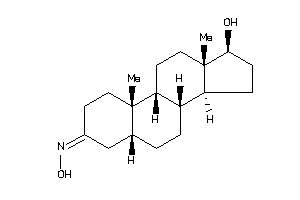
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5S,8R,9S,10S,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5S,8R,9S,10S,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.9 |
-5.3 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
12402554
-

-
12877346
-

-
13146629
-

-
13146631
-

-
4082983
-
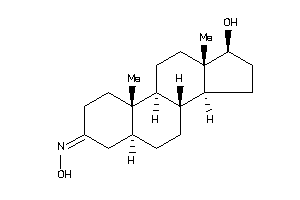
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,8R,9S,10R,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5R,8R,9S,10R,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.91 |
-6.27 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
12877346
-

-
13146629
-

-
13146631
-

-
4082983
-

-
12402551
-
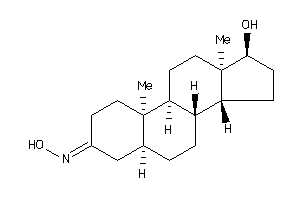
Draw
Identity
99%
90%
80%
70%
Popular Name:
(5R,8R,9S,10S,13R,14R,17S)-17-hydroxy-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydr
(5R,8R,9S,10S,13R,14R,17S)-17-hy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
4.71 |
-5.25 |
2 |
3 |
0 |
53 |
305.462 |
0 |
↓
|
|
|
|
Analogs
-
5276175
-
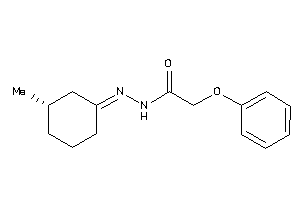
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
0.3 |
-15.16 |
1 |
4 |
0 |
51 |
260.337 |
4 |
↓
|
|
|
|
Analogs
-
12403626
-
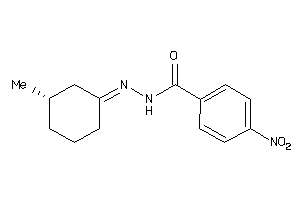
-
13138918
-
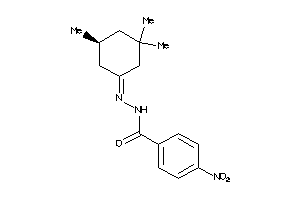
-
13138919
-
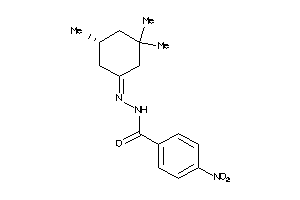
-
5783902
-
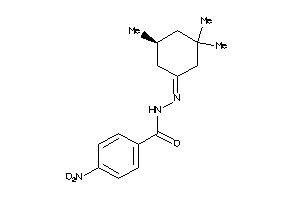
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
0.99 |
-12.44 |
1 |
6 |
0 |
87 |
275.308 |
3 |
↓
|
|
|
|
Analogs
-
13138918
-

-
13138919
-

-
5783902
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
0.96 |
-12.6 |
1 |
6 |
0 |
87 |
275.308 |
3 |
↓
|
|
|
|
Analogs
-
12403676
-
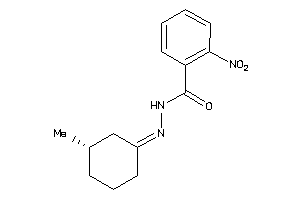
-
4957644
-
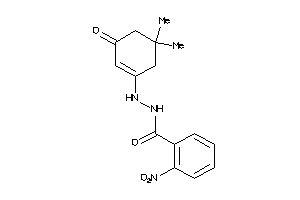
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
1.43 |
-22.65 |
1 |
6 |
0 |
87 |
275.308 |
3 |
↓
|
|
|
|
Analogs
-
4957644
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
0.82 |
-21.75 |
1 |
6 |
0 |
87 |
275.308 |
3 |
↓
|
|
|
|
Analogs
-
12403867
-
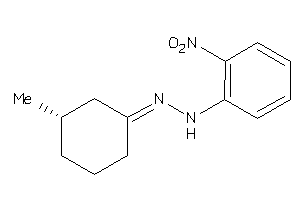
-
4599341
-
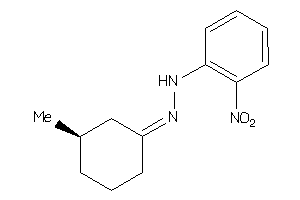
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
1.8 |
-8.26 |
1 |
5 |
0 |
70 |
247.298 |
3 |
↓
|
|
|
|
Analogs
-
4599341
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
1.77 |
-8.11 |
1 |
5 |
0 |
70 |
247.298 |
3 |
↓
|
|
|
|
Analogs
-
20569732
-
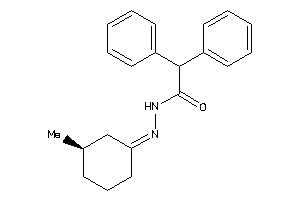
-
46115
-
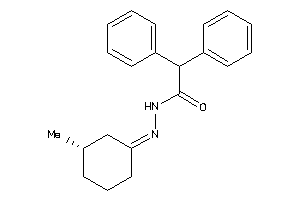
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
0.75 |
-12.04 |
1 |
3 |
0 |
41 |
320.436 |
4 |
↓
|
|
|
|
Analogs
-
12403982
-
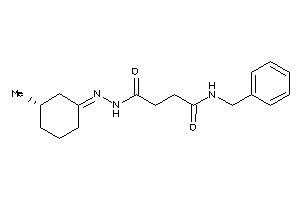
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
-1.96 |
-11.68 |
2 |
5 |
0 |
71 |
315.417 |
6 |
↓
|
|
|
|
Analogs
-
12403981
-
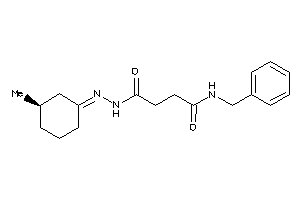
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
-1.99 |
-11.84 |
2 |
5 |
0 |
71 |
315.417 |
6 |
↓
|
|
|
|
Analogs
-
12403984
-
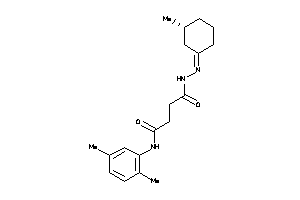
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
-0.28 |
-12.69 |
2 |
5 |
0 |
71 |
329.444 |
5 |
↓
|
|
|
|
Analogs
-
12403983
-
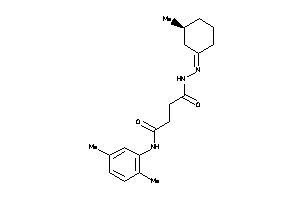
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
-0.29 |
-12.68 |
2 |
5 |
0 |
71 |
329.444 |
5 |
↓
|
|
|
|
Analogs
-
12404026
-
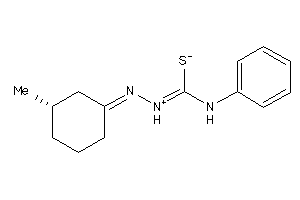
-
8685034
-
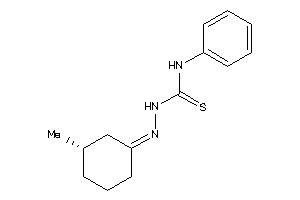
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
8.96 |
-9.46 |
2 |
3 |
0 |
36 |
261.394 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.86 |
8.55 |
-45.04 |
1 |
3 |
-1 |
37 |
260.386 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.86 |
8.88 |
-50.3 |
1 |
3 |
-1 |
37 |
260.386 |
3 |
↓
|
|
|
|
Analogs
-
12404025
-
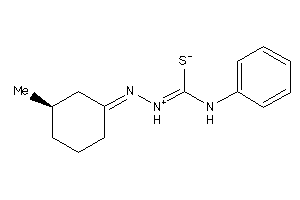
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
8.95 |
-9.46 |
2 |
3 |
0 |
36 |
261.394 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.86 |
8.57 |
-45.12 |
1 |
3 |
-1 |
37 |
260.386 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.91 |
9.87 |
-6.13 |
1 |
3 |
0 |
37 |
325.843 |
2 |
↓
|
|
|
|
Analogs
-
13013545
-
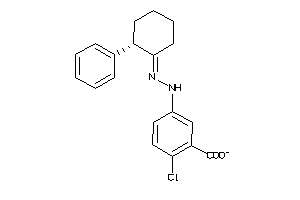
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.90 |
10.86 |
-50.75 |
1 |
4 |
-1 |
65 |
341.818 |
4 |
↓
|
|
|
|
Analogs
-
13013543
-
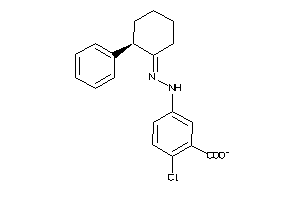
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.90 |
10.87 |
-50.55 |
1 |
4 |
-1 |
65 |
341.818 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
13.75 |
-28.66 |
1 |
8 |
0 |
105 |
496.365 |
6 |
↓
|
|
|
|
Analogs
-
7890663
-
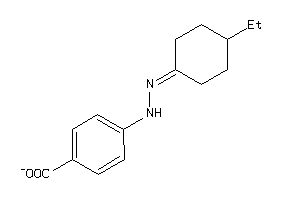
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.09 |
8.88 |
-53.62 |
1 |
4 |
-1 |
65 |
273.356 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.35 |
9.28 |
-50.16 |
1 |
4 |
-1 |
65 |
307.801 |
5 |
↓
|
|