|
|
|
|
|
|
|
|
Analogs
-
27738810
-
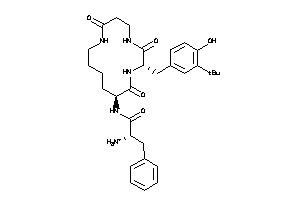
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
6.39 |
-57.88 |
8 |
10 |
1 |
164 |
622.831 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.41 |
6.13 |
-17.23 |
7 |
10 |
0 |
163 |
621.823 |
8 |
↓
|
|
|
|
Analogs
-
27738810
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
460 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.1 |
-58.69 |
8 |
10 |
1 |
164 |
622.831 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.41 |
6.82 |
-18.98 |
7 |
10 |
0 |
163 |
621.823 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36160452
-
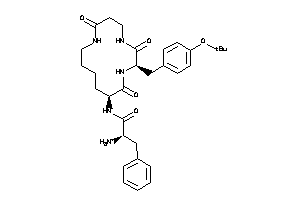
-
36160456
-
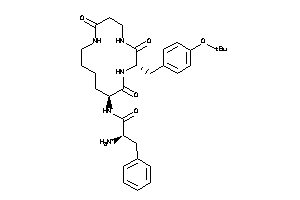
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7500 |
0.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
4.86 |
-53.63 |
7 |
10 |
1 |
153 |
566.723 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.60 |
4.59 |
-16.9 |
6 |
10 |
0 |
152 |
565.715 |
8 |
↓
|
|
|
|
Analogs
-
27738810
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTLR-1-E |
Motilin Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
4.38 |
-60.02 |
8 |
10 |
1 |
164 |
566.723 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.98 |
4.15 |
-19.58 |
7 |
10 |
0 |
163 |
565.715 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|