|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
6200 |
0.24 |
Binding ≤ 10μM
|
|
Q4U254-1-V |
Human Rhinovirus A Protease (cluster #1 Of 3), Viral |
Viruses |
6200 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.50 |
13.26 |
-43.98 |
1 |
4 |
-1 |
61 |
536.616 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
8.50 |
12.42 |
-9.5 |
2 |
4 |
0 |
59 |
537.624 |
4 |
↓
|
|
|
|
Analogs
-
3830631
-
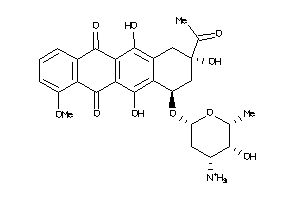
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
-11.06 |
-58.55 |
8 |
12 |
1 |
207 |
544.533 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF-1-E |
Cathepsin F (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
|
D2K2A8-1-V |
Hepatitis C Virus NS4A Protein (cluster #1 Of 1), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.12 |
-12.14 |
5 |
10 |
0 |
151 |
519.687 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB-1-E |
Cathepsin B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
CATF-1-E |
Cathepsin F (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.23 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.28 |
Binding ≤ 10μM
|
|
CATL1-1-E |
Cathepsin L (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
760 |
0.23 |
Binding ≤ 10μM
|
|
CATL2-1-E |
Cathepsin L2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.27 |
Binding ≤ 10μM
|
|
CATS-2-E |
Cathepsin S (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.26 |
Binding ≤ 10μM
|
|
CMA1-1-E |
Mast Cell Protease 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.28 |
Binding ≤ 10μM
|
|
Z50643-1-O |
Hepatitis C Virus (cluster #1 Of 2), Other |
Other |
14 |
0.30 |
Binding ≤ 10μM
|
|
Z50643-5-O |
Hepatitis C Virus (cluster #5 Of 5), Other |
Other |
200 |
0.25 |
Functional ≤ 10μM |
|
A3EZI9-2-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #2 Of 3), Viral |
Viruses |
14 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.18 |
-13.07 |
5 |
10 |
0 |
151 |
519.687 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
200 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
-1.39 |
-43.83 |
1 |
3 |
-1 |
56 |
239.048 |
1 |
↓
|
|
|
|
Analogs
-
26474911
-
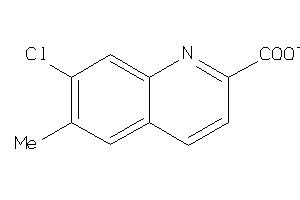
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
400 |
0.69 |
Binding ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
10000 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
-0.78 |
-43.98 |
1 |
3 |
-1 |
56 |
194.597 |
1 |
↓
|
|
|
|
Analogs
-
32003100
-
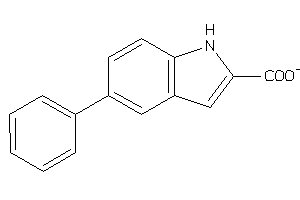
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1200 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
-0.31 |
-47.78 |
1 |
3 |
-1 |
56 |
174.179 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
2900 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
5.05 |
-42.82 |
1 |
3 |
-1 |
56 |
178.142 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 59 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
300 |
0.70 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
5.51 |
-42.76 |
1 |
3 |
-1 |
56 |
194.597 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 62 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
4000 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
3.28 |
-47.52 |
1 |
4 |
-1 |
65 |
190.178 |
2 |
↓
|
|
|
|
Analogs
-
34567204
-
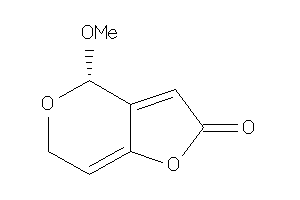
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3800 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
0.94 |
-12.15 |
1 |
4 |
0 |
60 |
154.121 |
0 |
↓
|
|
|
|
Analogs
-
34567204
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3800 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
0.94 |
-12.15 |
1 |
4 |
0 |
60 |
154.121 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
34364656
-
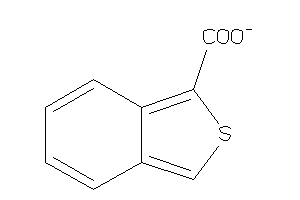
Draw
Identity
99%
90%
80%
70%
Vendors
And 60 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1700 |
0.67 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
-0.26 |
-52.07 |
0 |
2 |
-1 |
40 |
177.204 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
800 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
-0.78 |
-47.19 |
1 |
3 |
-1 |
56 |
194.597 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
6500 |
0.18 |
Binding ≤ 10μM
|
|
Q4U254-1-V |
Human Rhinovirus A Protease (cluster #1 Of 3), Viral |
Viruses |
6500 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.71 |
13.85 |
-45.68 |
1 |
6 |
-1 |
87 |
631.344 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.71 |
13.84 |
-13.68 |
2 |
6 |
0 |
85 |
632.352 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3200 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
9.51 |
-43.41 |
0 |
5 |
-1 |
82 |
407.905 |
4 |
↓
|
|
|
|
Analogs
-
21803838
-
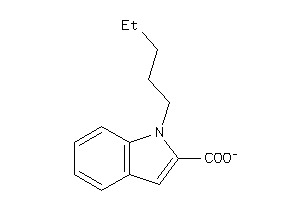
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
8100 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
1.01 |
-48.72 |
0 |
3 |
-1 |
45 |
174.179 |
1 |
↓
|
|
|
|
Analogs
-
6645691
-
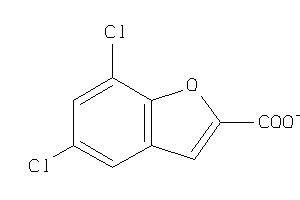
-
37034694
-
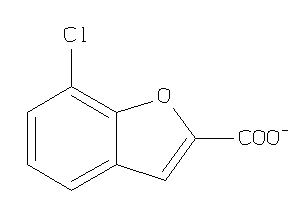
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
2200 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
0.84 |
-46.98 |
0 |
3 |
-1 |
53 |
195.581 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
411 |
0.32 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.42 |
Binding ≤ 10μM
|
|
BGLR-2-E |
Beta-glucuronidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
CALM-2-E |
Calmodulin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
EBP-1-E |
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
|
LYSC1-1-E |
Lysozyme C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.25 |
Binding ≤ 10μM
|
|
MDR1-2-E |
P-glycoprotein 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.26 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.39 |
Binding ≤ 10μM
|
|
DRD2-14-E |
Dopamine D2 Receptor (cluster #14 Of 24), Eukaryotic |
Eukaryotes |
2 |
0.43 |
Binding ≤ 10μM
|
|
ERG2-2-F |
C-8 Sterol Isomerase (cluster #2 Of 2), Fungal |
Fungi |
500 |
0.32 |
Binding ≤ 10μM
|
|
PDR5-1-F |
Pleiotropic ABC Efflux Transporter Of Multiple Drugs (cluster #1 Of 1), Fungal |
Fungi |
1400 |
0.29 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
350 |
0.32 |
Binding ≤ 10μM
|
|
Z100748-2-O |
Neutrophils (cluster #2 Of 2), Other |
Other |
6600 |
0.26 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
3150 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6000 |
0.26 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
6600 |
0.26 |
Functional ≤ 10μM
|
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
700 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
11.43 |
-38.84 |
1 |
3 |
1 |
13 |
408.513 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.14 |
9.06 |
-5.72 |
0 |
3 |
0 |
11 |
407.505 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.14 |
11.32 |
-44.3 |
1 |
3 |
1 |
13 |
408.513 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1400 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
-0.78 |
-46.87 |
1 |
3 |
-1 |
56 |
194.597 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ERG1-3-E |
Squalene Monooxygenase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
630 |
0.19 |
Binding ≤ 10μM
|
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1890 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
-16.67 |
-32.09 |
11 |
18 |
0 |
310 |
636.471 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3000 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
8.8 |
-49.57 |
0 |
3 |
-1 |
57 |
259.668 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
4000 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.08 |
2.12 |
-51.78 |
0 |
3 |
-1 |
57 |
243.213 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1000 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
1.44 |
-55.54 |
0 |
4 |
-1 |
66 |
255.249 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 40 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1600 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
1.71 |
-56.73 |
0 |
3 |
-1 |
57 |
239.25 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1000 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
-1.4 |
-46.83 |
1 |
3 |
-1 |
56 |
239.048 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
8000 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
4.9 |
-50.84 |
0 |
3 |
-1 |
49 |
207.23 |
2 |
↓
|
|
|
|
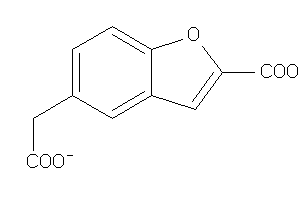
Analogs
-
16431115
-
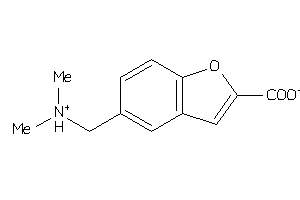
-
34525766
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 65 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
6400 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
0.99 |
-52.2 |
0 |
3 |
-1 |
53 |
161.136 |
1 |
↓
|
|
|
|
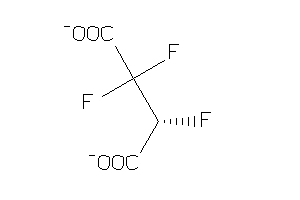
Analogs
-
15442369
-
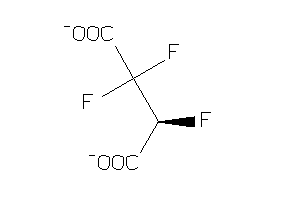
-
15442370
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-3-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #3 Of 3), Viral |
Viruses |
5200 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.09 |
1.99 |
-94.97 |
0 |
4 |
-2 |
80 |
188.032 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
1600 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
-0.54 |
-54.3 |
1 |
4 |
-1 |
77 |
241.222 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
690 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
3.81 |
-7.66 |
2 |
3 |
0 |
49 |
259.227 |
3 |
↓
|
|
|
|
|