|
|
Analogs
-
44126678
-
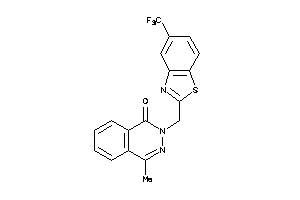
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
0.96 |
-59.14 |
0 |
6 |
-1 |
87 |
418.376 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.35 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
36 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
5.35 |
-28.08 |
3 |
8 |
1 |
108 |
418.429 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.88 |
-3.15 |
-47.34 |
3 |
8 |
1 |
107 |
418.429 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
36 |
0.33 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
6.43 |
-33.11 |
3 |
8 |
1 |
108 |
432.456 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.54 |
-2.76 |
-46.75 |
3 |
8 |
1 |
107 |
432.456 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.35 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
21 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
-2.8 |
-30.78 |
2 |
8 |
0 |
106 |
417.421 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.88 |
-3.12 |
-46.15 |
3 |
8 |
1 |
107 |
418.429 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.40 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
11 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
5.93 |
-30.4 |
3 |
7 |
1 |
99 |
388.403 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.90 |
-3.24 |
-44.77 |
3 |
7 |
1 |
98 |
388.403 |
4 |
↓
|
|
|
|
Analogs
-
2574862
-
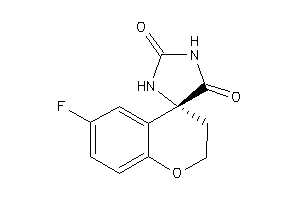
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
0.48 |
-8.57 |
2 |
5 |
0 |
67 |
236.202 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.05 |
-2.11 |
-43.04 |
1 |
5 |
-1 |
74 |
235.194 |
0 |
↓
|
|
|
|
Analogs
-
1492564
-
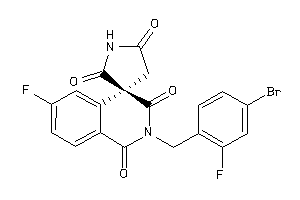
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1A1-1-E |
Aldehyde Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.37 |
Binding ≤ 10μM
|
|
AK1BA-1-E |
Aldo-keto Reductase Family 1 Member B10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.31 |
Binding ≤ 10μM
|
|
ALDR-2-E |
Aldose Reductase (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
14 |
0.39 |
Binding ≤ 10μM
|
|
PPBT-1-E |
Alkaline Phosphatase Tissue-nonspecific (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
-0.4 |
-10.22 |
1 |
6 |
0 |
83 |
449.207 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
10.02 |
-52.28 |
0 |
4 |
-1 |
53 |
356.345 |
6 |
↓
|
|
|
|
|