|
|
Analogs
-
1802
-
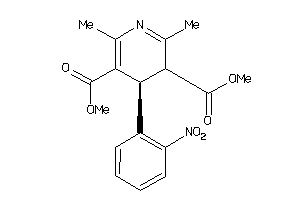
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2890 |
0.31 |
Binding ≤ 10μM
|
|
AA2AR-4-E |
Adenosine A2a Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8290 |
0.28 |
Binding ≤ 10μM
|
|
CAC1A-1-E |
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4300 |
0.30 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Functional ≤ 10μM |
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
ADME/T ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 1), Other |
Other |
1 |
0.50 |
Binding ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
20 |
0.43 |
Functional ≤ 10μM
|
|
Z102372-1-O |
Aorta (cluster #1 Of 1), Other |
Other |
9 |
0.45 |
Functional ≤ 10μM |
|
Z102380-2-O |
Ileum (cluster #2 Of 3), Other |
Other |
15 |
0.44 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
4 |
0.47 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
7 |
0.46 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
9 |
0.45 |
Functional ≤ 10μM
|
|
Z80051-1-O |
C6-BU-1 (cluster #1 Of 1), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
8.22 |
-42.09 |
0 |
8 |
-1 |
117 |
345.331 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.32 |
7.84 |
-40.28 |
0 |
8 |
-1 |
117 |
345.331 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
90 |
0.29 |
Binding ≤ 10μM
|
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.29 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.29 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.31 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.31 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
9000 |
0.21 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
12.82 |
-48.51 |
1 |
5 |
1 |
35 |
469.552 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.97 |
10.52 |
-9.52 |
0 |
5 |
0 |
34 |
468.544 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.97 |
12.37 |
-49.34 |
1 |
5 |
1 |
35 |
469.552 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.29 |
Binding ≤ 10μM
|
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.25 |
Binding ≤ 10μM
|
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.28 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.33 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3630 |
0.25 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9480 |
0.23 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2410 |
0.26 |
Binding ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.30 |
Binding ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.30 |
Binding ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.29 |
Binding ≤ 10μM
|
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Functional ≤ 10μM
|
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Functional ≤ 10μM
|
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Functional ≤ 10μM
|
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.32 |
Functional ≤ 10μM
|
|
SCN1A-1-E |
Sodium Channel Protein Type I Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.31 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
420 |
0.30 |
Functional ≤ 10μM
|
|
SCN3A-1-E |
Sodium Channel Protein Type III Alpha Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.31 |
Functional ≤ 10μM
|
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.32 |
Functional ≤ 10μM
|
|
SCN8A-1-E |
Sodium Channel Protein Type VIII Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
290 |
0.31 |
Functional ≤ 10μM
|
|
DRD2-17-E |
Dopamine D2 Receptor (cluster #17 Of 24), Eukaryotic |
Eukaryotes |
228 |
0.31 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
3981 |
0.25 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7300 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.09 |
14.99 |
-46.52 |
1 |
2 |
1 |
8 |
405.512 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.09 |
14.54 |
-45.46 |
1 |
2 |
1 |
8 |
405.512 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.09 |
12.7 |
-5.38 |
0 |
2 |
0 |
6 |
404.504 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
150 |
0.27 |
Binding ≤ 10μM
|
|
CAC1B-2-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.27 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
15.94 |
-10.06 |
1 |
4 |
0 |
36 |
461.609 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.79 |
17.52 |
-45.14 |
2 |
4 |
1 |
37 |
462.617 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
260 |
0.26 |
Binding ≤ 10μM
|
|
CAC1B-2-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.26 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
260 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.16 |
15 |
-9.39 |
0 |
4 |
0 |
27 |
461.609 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.16 |
17.19 |
-38.3 |
1 |
4 |
1 |
28 |
462.617 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.16 |
16.58 |
-35.98 |
1 |
4 |
1 |
28 |
462.617 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
110 |
0.28 |
Binding ≤ 10μM
|
|
CAC1B-2-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.28 |
Binding ≤ 10μM
|
|
CACB1-1-E |
Voltage-dependent L-type Calcium Channel Subunit Beta-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.19 |
16.53 |
-9.31 |
0 |
3 |
0 |
24 |
460.621 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.19 |
18.34 |
-45.26 |
1 |
3 |
1 |
25 |
461.629 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1A-1-E |
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3860 |
0.33 |
Functional ≤ 10μM |
|
P97706-1-E |
Sodium Channel Protein Type VI Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.32 |
Functional ≤ 10μM
|
|
SCN2A-1-E |
Sodium Channel Protein Type II Alpha Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2550 |
0.34 |
Functional ≤ 10μM |
|
SCN5A-1-E |
Sodium Channel Protein Type V Alpha Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5300 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
7000 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
-3.27 |
-37.36 |
3 |
5 |
1 |
59 |
373.695 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.30 |
-3.11 |
-94.34 |
4 |
5 |
2 |
60 |
374.703 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.30 |
-3.01 |
-90.06 |
4 |
5 |
2 |
60 |
374.703 |
2 |
↓
|
|