|
|
Analogs
-
33799539
-
-
44608728
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTSI-1-B |
CpG DNA Methylase (cluster #1 Of 2), Bacterial |
Bacteria |
30 |
0.39 |
Binding ≤ 10μM
|
|
A4-1-E |
Beta Amyloid A4 Protein (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
800 |
0.32 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2410 |
0.29 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
3480 |
0.28 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6850 |
0.27 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5090 |
0.27 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
380 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4970 |
0.28 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9460 |
0.26 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
9940 |
0.26 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
9300 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4050 |
0.28 |
Binding ≤ 10μM
|
|
DHB3-1-E |
Estradiol 17-beta-dehydrogenase 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.26 |
Binding ≤ 10μM
|
|
KPCE-5-E |
Protein Kinase C Epsilon (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
8810 |
0.26 |
Binding ≤ 10μM
|
|
LGUL-2-E |
Glyoxalase I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
LOX5-4-E |
Arachidonate 5-lipoxygenase (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8000 |
0.26 |
Binding ≤ 10μM
|
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.26 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
PGH1-6-E |
Cyclooxygenase-1 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
8800 |
0.26 |
Binding ≤ 10μM
|
|
Q8HY88-1-E |
Potassium Channel Subfamily K Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
930 |
0.31 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4300 |
0.28 |
ADME/T ≤ 10μM
|
|
Z103192-1-O |
Trypanosoma Evansi (cluster #1 Of 2), Other |
Other |
2000 |
0.30 |
Functional ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
4800 |
0.28 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
4210 |
0.28 |
Functional ≤ 10μM
|
|
Z50515-1-O |
Human Herpesvirus 2 (cluster #1 Of 2), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50518-1-O |
Human Herpesvirus 4 (cluster #1 Of 5), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50600-2-O |
Vaccinia Virus (cluster #2 Of 2), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50602-1-O |
Human Herpesvirus 1 (cluster #1 Of 5), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
37 |
0.39 |
Functional ≤ 10μM
|
|
Z50651-2-O |
Vesicular Stomatitis Virus (cluster #2 Of 2), Other |
Other |
10 |
0.41 |
Functional ≤ 10μM
|
|
Z50658-4-O |
Human Immunodeficiency Virus 2 (cluster #4 Of 4), Other |
Other |
37 |
0.39 |
Functional ≤ 10μM
|
|
Z80186-2-O |
K562 (Erythroleukemia Cells) (cluster #2 Of 11), Other |
Other |
6810 |
0.27 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5580 |
0.27 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
9700 |
0.26 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7700 |
0.27 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
8300 |
0.26 |
Functional ≤ 10μM
|
|
Z80548-3-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #3 Of 5), Other |
Other |
1210 |
0.31 |
Functional ≤ 10μM |
|
Z80612-1-O |
2008 (Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
8500 |
0.26 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6500 |
0.27 |
Functional ≤ 10μM
|
|
Z81252-1-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #1 Of 11), Other |
Other |
7600 |
0.27 |
Functional ≤ 10μM
|
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
9000 |
0.26 |
ADME/T ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 4), Other |
Other |
8700 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
7.14 |
-22.28 |
2 |
6 |
0 |
93 |
368.385 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
5.47 |
-58.07 |
2 |
6 |
-1 |
99 |
367.377 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
140 |
0.37 |
Binding ≤ 10μM
|
|
CYNT-1-B |
Carbonic Anhydrase (cluster #1 Of 3), Bacterial |
Bacteria |
713 |
0.33 |
Binding ≤ 10μM
|
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
34 |
0.40 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
690 |
0.33 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
18 |
0.42 |
Binding ≤ 10μM |
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.38 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
689 |
0.33 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
45 |
0.40 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Binding ≤ 10μM |
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
794 |
0.33 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
93 |
0.38 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.38 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2170 |
0.30 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM |
|
COX2-1-E |
Cytochrome C Oxidase Subunit 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.39 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
810 |
0.33 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9730 |
0.27 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9 |
0.43 |
Binding ≤ 10μM
|
|
Q8HZR1-1-E |
Cyclooxygenase-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5570 |
0.28 |
Binding ≤ 10μM
|
|
Q8SPQ9-2-E |
Cyclooxygenase-2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.33 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Functional ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
ADME/T ≤ 10μM
|
|
CAN-1-F |
Carbonic Anhydrase (cluster #1 Of 3), Fungal |
Fungi |
108 |
0.38 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
21 |
0.41 |
Binding ≤ 10μM
|
|
Z100741-1-O |
MC9 (Mast Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6670 |
0.28 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
5000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
5.33 |
-11.94 |
2 |
5 |
0 |
78 |
381.379 |
4 |
↓
|
|
|
|
Analogs
-
12353732
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
169 |
0.56 |
Binding ≤ 10μM
|
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2210 |
0.47 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
950 |
0.50 |
Binding ≤ 10μM
|
|
CAH13-7-E |
Carbonic Anhydrase XIII (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
4090 |
0.44 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9360 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
2770 |
0.46 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9090 |
0.42 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4470 |
0.44 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
4750 |
0.44 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4640 |
0.44 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8070 |
0.42 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4350 |
0.44 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
810 |
0.50 |
Binding ≤ 10μM
|
|
ESR1-5-E |
Estrogen Receptor Alpha (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
785 |
0.50 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.50 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3490 |
0.45 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2840 |
0.46 |
Functional ≤ 10μM
|
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
6918 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
5000 |
0.44 |
Functional ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 9), Other |
Other |
4000 |
0.44 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
0.79 |
-8.46 |
3 |
3 |
0 |
61 |
228.247 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
190 |
0.59 |
Binding ≤ 10μM
|
|
CYNT-1-B |
Carbonic Anhydrase (cluster #1 Of 3), Bacterial |
Bacteria |
2010 |
0.50 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
872 |
0.53 |
Binding ≤ 10μM
|
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
431 |
0.56 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
79 |
0.62 |
Binding ≤ 10μM
|
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
374 |
0.56 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
56 |
0.63 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
23 |
0.67 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
345 |
0.57 |
Binding ≤ 10μM |
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
95 |
0.61 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
9 |
0.70 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
380 |
0.56 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
81 |
0.62 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
91 |
0.62 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
79 |
0.62 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.72 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
501 |
0.55 |
Binding ≤ 10μM
|
|
CAH1-2-E |
Carbonic Anhydrase I (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.52 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.65 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
380 |
0.56 |
Functional ≤ 10μM
|
|
CAN-1-F |
Carbonic Anhydrase (cluster #1 Of 3), Fungal |
Fungi |
103 |
0.61 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
910 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
-4.57 |
-13.7 |
4 |
6 |
0 |
120 |
305.164 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
170 |
0.68 |
Binding ≤ 10μM
|
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
660 |
0.62 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
562 |
0.63 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
781 |
0.61 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
21 |
0.77 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
132 |
0.69 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
780 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM |
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
19 |
0.77 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
43 |
0.74 |
Binding ≤ 10μM |
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
65 |
0.72 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.90 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
36 |
0.74 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
65 |
0.72 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
62 |
0.72 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Binding ≤ 10μM |
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
27 |
0.76 |
Binding ≤ 10μM
|
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.73 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.79 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
36 |
0.74 |
Functional ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
119 |
0.69 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
484 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.15 |
-5.18 |
-53.63 |
3 |
7 |
0 |
109 |
237.286 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
-0.15 |
-4.78 |
-45.79 |
3 |
7 |
0 |
109 |
237.286 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.15 |
-3.64 |
-23.75 |
2 |
7 |
0 |
106 |
236.278 |
1 |
↓
|
|
|
|
Analogs
-
4217364
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
60 |
0.78 |
Binding ≤ 10μM
|
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
9 |
0.87 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
12 |
0.85 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
481 |
0.68 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAH1-10-E |
Carbonic Anhydrase I (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
900 |
0.65 |
Binding ≤ 10μM
|
|
CAH10-2-E |
Carbonic Anhydrase-related Protein 10 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH11-2-E |
Carbonic Anhydrase-related Protein 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM |
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
490 |
0.68 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH2-11-E |
Carbonic Anhydrase II (cluster #11 Of 15), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
54 |
0.78 |
Binding ≤ 10μM |
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH8-2-E |
Carbonic Anhydrase-related Protein 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
1000 |
0.65 |
Binding ≤ 10μM
|
|
Q2PCB5-1-E |
Carbonic Anhydrase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5720 |
0.56 |
Binding ≤ 10μM |
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.92 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Functional ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
83 |
0.76 |
Binding ≤ 10μM
|
|
Q3I4V7-2-F |
Carbonic Anhydrase 2 (cluster #2 Of 4), Fungal |
Fungi |
10 |
0.86 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
40 |
0.80 |
Binding ≤ 10μM
|
|
Q6FTL6-1-F |
Carbonic Anhydrase (cluster #1 Of 1), Fungal |
Fungi |
11 |
0.86 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.36 |
-19.16 |
3 |
7 |
0 |
115 |
222.251 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.15 |
-3.88 |
-51.46 |
2 |
7 |
-1 |
113 |
221.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
3930 |
0.69 |
Binding ≤ 10μM
|
|
CYNT-1-B |
Carbonic Anhydrase (cluster #1 Of 3), Bacterial |
Bacteria |
454 |
0.81 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
7110 |
0.66 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
9840 |
0.64 |
Binding ≤ 10μM
|
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
364 |
0.82 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
37 |
0.95 |
Binding ≤ 10μM |
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
32 |
0.95 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5400 |
0.67 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
9610 |
0.64 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
988 |
0.76 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
3000 |
0.70 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
1360 |
0.75 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
3650 |
0.69 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
941 |
0.77 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
70 |
0.91 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
294 |
0.83 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Functional ≤ 10μM
|
|
CAH9-2-E |
Carbonic Anhydrase IX (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
294 |
0.83 |
Functional ≤ 10μM
|
|
Q3I4V7-3-F |
Carbonic Anhydrase 2 (cluster #3 Of 4), Fungal |
Fungi |
770 |
0.78 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
7630 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.29 |
-2.67 |
-10.63 |
4 |
4 |
0 |
86 |
172.209 |
1 |
↓
|
|
|
|
Analogs
-
21984495
-
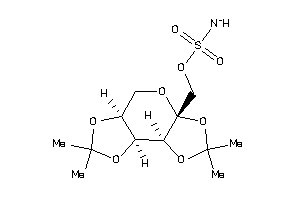
-
22061272
-
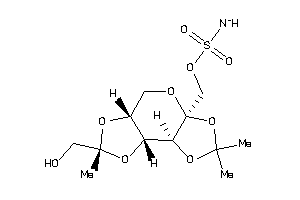
-
22061274
-
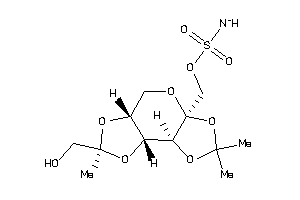
-
23586802
-
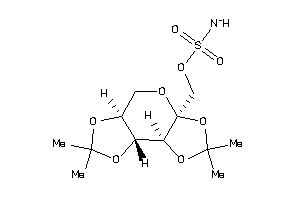
-
23586806
-
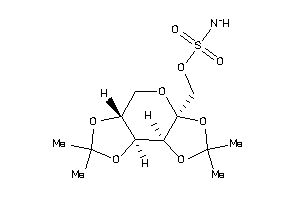
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-2-A |
Carbonic Anhydrase (cluster #2 Of 2), Archaea |
Archaea |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CYNT-3-B |
Carbonic Anhydrase (cluster #3 Of 3), Bacterial |
Bacteria |
474 |
0.40 |
Binding ≤ 10μM
|
|
P96878-2-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #2 Of 2), Bacterial |
Bacteria |
3020 |
0.35 |
Binding ≤ 10μM
|
|
Y1284-2-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #2 Of 2), Bacterial |
Bacteria |
612 |
0.40 |
Binding ≤ 10μM
|
|
B5SU02-4-E |
Alpha Carbonic Anhydrase (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
29 |
0.48 |
Binding ≤ 10μM
|
|
C0IX24-3-E |
Carbonic Anhydrase (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
367 |
0.41 |
Binding ≤ 10μM
|
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3800 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-3-E |
Carbonic Anhydrase XIII (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
47 |
0.47 |
Binding ≤ 10μM
|
|
CAH14-5-E |
Carbonic Anhydrase XIV (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1460 |
0.37 |
Binding ≤ 10μM
|
|
CAH15-5-E |
Carbonic Anhydrase 15 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
CAH2-10-E |
Carbonic Anhydrase II (cluster #10 Of 15), Eukaryotic |
Eukaryotes |
8900 |
0.32 |
Binding ≤ 10μM
|
|
CAH3-4-E |
Carbonic Anhydrase III (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH4-7-E |
Carbonic Anhydrase IV (cluster #7 Of 16), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Binding ≤ 10μM
|
|
CAH5A-3-E |
Carbonic Anhydrase VA (cluster #3 Of 10), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH5B-3-E |
Carbonic Anhydrase VB (cluster #3 Of 9), Eukaryotic |
Eukaryotes |
30 |
0.48 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8890 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-5-E |
Carbonic Anhydrase VII (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
58 |
0.46 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Functional ≤ 10μM
|
|
CAH2-2-E |
Carbonic Anhydrase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
CAH4-2-E |
Carbonic Anhydrase IV (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
54 |
0.46 |
Functional ≤ 10μM
|
|
CAN-2-F |
Carbonic Anhydrase (cluster #2 Of 3), Fungal |
Fungi |
110 |
0.44 |
Binding ≤ 10μM
|
|
Q3I4V7-4-F |
Carbonic Anhydrase 2 (cluster #4 Of 4), Fungal |
Fungi |
370 |
0.41 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
1110 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
-1.52 |
-44.79 |
1 |
9 |
-1 |
113 |
338.358 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.16 |
-2 |
-12.26 |
2 |
9 |
0 |
116 |
339.366 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
2090 |
0.80 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
105 |
0.98 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
361 |
0.90 |
Binding ≤ 10μM
|
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
8600 |
0.71 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
34 |
1.05 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
79 |
0.99 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
280 |
0.92 |
Binding ≤ 10μM
|
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
48 |
1.02 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
7400 |
0.72 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
540 |
0.88 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
2300 |
0.79 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
2150 |
0.79 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
798 |
0.85 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
5 |
1.16 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
41 |
1.03 |
Binding ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
447 |
0.89 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
832 |
0.85 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.88 |
-5.22 |
-12.61 |
4 |
6 |
0 |
112 |
180.214 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.88 |
-4.74 |
-44.86 |
3 |
6 |
-1 |
109 |
179.206 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH10-1-E |
Carbonic Anhydrase-related Protein 10 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH11-1-E |
Carbonic Anhydrase-related Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
4200 |
0.58 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH8-1-E |
Carbonic Anhydrase-related Protein 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.59 |
0.17 |
-11.69 |
2 |
6 |
0 |
106 |
202.191 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH10-1-E |
Carbonic Anhydrase-related Protein 10 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH11-1-E |
Carbonic Anhydrase-related Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH8-1-E |
Carbonic Anhydrase-related Protein 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
146 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
1.38 |
-10.64 |
1 |
6 |
0 |
92 |
216.218 |
3 |
↓
|
|
|
|
Analogs
-
39332864
-
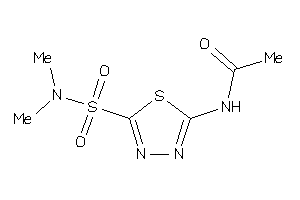
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-10-E |
Carbonic Anhydrase I (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH10-2-E |
Carbonic Anhydrase-related Protein 10 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH11-2-E |
Carbonic Anhydrase-related Protein 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH2-11-E |
Carbonic Anhydrase II (cluster #11 Of 15), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH8-2-E |
Carbonic Anhydrase-related Protein 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
258 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.77 |
-3.07 |
-18.21 |
2 |
7 |
0 |
101 |
236.278 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.77 |
-3.59 |
-51.44 |
1 |
7 |
-1 |
103 |
235.27 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH10-1-E |
Carbonic Anhydrase-related Protein 10 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH11-1-E |
Carbonic Anhydrase-related Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH8-1-E |
Carbonic Anhydrase-related Protein 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
584 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
1.19 |
-8.12 |
1 |
3 |
0 |
46 |
205.666 |
2 |
↓
|
|
|
|
Analogs
-
643114
-
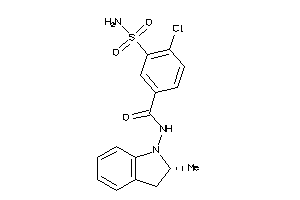
-
3644963
-
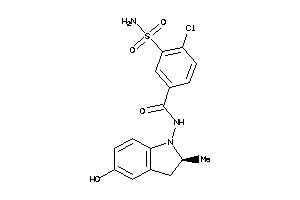
-
3644965
-
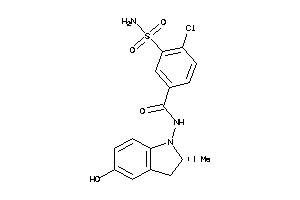
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
13 |
0.46 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4950 |
0.31 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
213 |
0.39 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
890 |
0.35 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
274 |
0.38 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1606 |
0.34 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
36 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
3.92 |
-19.14 |
3 |
6 |
0 |
93 |
365.842 |
3 |
↓
|
|
|
|
Analogs
-
3644963
-

-
3644965
-

-
601305
-
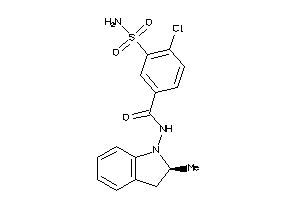
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
10000 |
0.29 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
13 |
0.46 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4950 |
0.31 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
213 |
0.39 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
890 |
0.35 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
274 |
0.38 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1606 |
0.34 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
36 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
3.93 |
-19.38 |
3 |
6 |
0 |
93 |
365.842 |
3 |
↓
|
|
|
|
Analogs
-
26387469
-
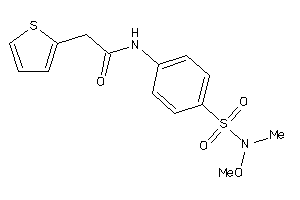
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
61 |
0.53 |
Binding ≤ 10μM |
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
125 |
0.51 |
Binding ≤ 10μM |
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM |
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
7 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.61 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
1.28 |
-17.29 |
3 |
5 |
0 |
89 |
296.373 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
161 |
0.48 |
Binding ≤ 10μM |
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
437 |
0.45 |
Binding ≤ 10μM |
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
390 |
0.45 |
Binding ≤ 10μM |
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
7 |
0.57 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
1.38 |
-15.26 |
3 |
5 |
0 |
89 |
314.363 |
4 |
↓
|
|
|
|
Analogs
-
29562604
-
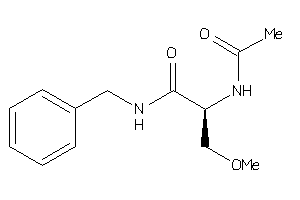
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
362 |
0.50 |
Binding ≤ 10μM
|
|
CAH12-6-E |
Carbonic Anhydrase XII (cluster #6 Of 9), Eukaryotic |
Eukaryotes |
3713 |
0.42 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1210 |
0.46 |
Binding ≤ 10μM
|
|
CAH14-8-E |
Carbonic Anhydrase XIV (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
473 |
0.49 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
461 |
0.49 |
Binding ≤ 10μM
|
|
CAH2-14-E |
Carbonic Anhydrase II (cluster #14 Of 15), Eukaryotic |
Eukaryotes |
331 |
0.50 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
374 |
0.50 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
525 |
0.49 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
4565 |
0.42 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
341 |
0.50 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
412 |
0.50 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4446 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-7-E |
Carbonic Anhydrase IX (cluster #7 Of 11), Eukaryotic |
Eukaryotes |
353 |
0.50 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-2.84 |
-9.35 |
2 |
5 |
0 |
67 |
250.298 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-8-E |
Carbonic Anhydrase I (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
50 |
1.28 |
Binding ≤ 10μM
|
|
CAH13-4-E |
Carbonic Anhydrase XIII (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
27 |
1.32 |
Binding ≤ 10μM
|
|
CAH2-8-E |
Carbonic Anhydrase II (cluster #8 Of 15), Eukaryotic |
Eukaryotes |
91 |
1.23 |
Binding ≤ 10μM
|
|
CAH7-5-E |
Carbonic Anhydrase VII (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
36 |
1.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
-3.19 |
-7.94 |
2 |
3 |
0 |
60 |
149.093 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
876 |
0.61 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
210 |
0.67 |
Binding ≤ 10μM
|
|
AOFB-4-E |
Monoamine Oxidase B (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.55 |
Binding ≤ 10μM |
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
259 |
0.66 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
645 |
0.62 |
Binding ≤ 10μM
|
|
CAH1-12-E |
Carbonic Anhydrase I (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
56 |
0.73 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
430 |
0.64 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5250 |
0.53 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
634 |
0.62 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
6800 |
0.52 |
Binding ≤ 10μM
|
|
CAH4-14-E |
Carbonic Anhydrase IV (cluster #14 Of 16), Eukaryotic |
Eukaryotes |
8590 |
0.51 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
21 |
0.77 |
Binding ≤ 10μM
|
|
CAH5B-9-E |
Carbonic Anhydrase VB (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
6033 |
0.52 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
89 |
0.71 |
Binding ≤ 10μM
|
|
CAH7-8-E |
Carbonic Anhydrase VII (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
117 |
0.69 |
Binding ≤ 10μM
|
|
CAH9-9-E |
Carbonic Anhydrase IX (cluster #9 Of 11), Eukaryotic |
Eukaryotes |
5 |
0.83 |
Binding ≤ 10μM
|
|
CAN-1-F |
Carbonic Anhydrase (cluster #1 Of 3), Fungal |
Fungi |
106 |
0.70 |
Binding ≤ 10μM
|
|
Q3I4V7-3-F |
Carbonic Anhydrase 2 (cluster #3 Of 4), Fungal |
Fungi |
970 |
0.60 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
942 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
-0.51 |
-18.88 |
2 |
5 |
0 |
86 |
212.23 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-5-E |
Carbonic Anhydrase I (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH10-1-E |
Carbonic Anhydrase-related Protein 10 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH11-1-E |
Carbonic Anhydrase-related Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH2-6-E |
Carbonic Anhydrase II (cluster #6 Of 15), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH6-7-E |
Carbonic Anhydrase VI (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH8-1-E |
Carbonic Anhydrase-related Protein 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.31 |
-0.03 |
-8.88 |
2 |
3 |
0 |
60 |
191.639 |
1 |
↓
|
|
|
|
Analogs
-
32150186
-
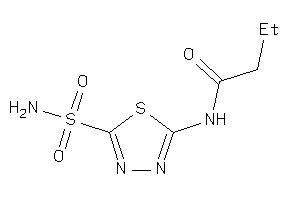
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-10-E |
Carbonic Anhydrase I (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH10-2-E |
Carbonic Anhydrase-related Protein 10 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH11-2-E |
Carbonic Anhydrase-related Protein 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH2-11-E |
Carbonic Anhydrase II (cluster #11 Of 15), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH8-2-E |
Carbonic Anhydrase-related Protein 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.40 |
-2.86 |
-52.35 |
3 |
9 |
-1 |
155 |
279.279 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.40 |
-2.37 |
-92.86 |
2 |
9 |
-2 |
153 |
278.271 |
5 |
↓
|
|
|
|
Analogs
-
57278
-
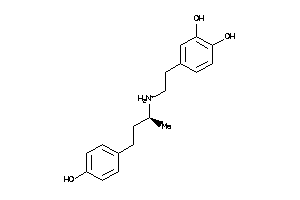
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9530 |
0.32 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
470 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
3.38 |
-54.79 |
5 |
4 |
1 |
77 |
302.394 |
7 |
↓
|
|
|
|
Analogs
-
3911
-
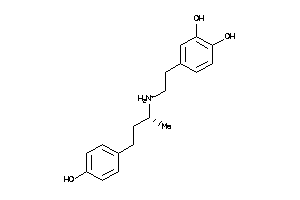
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1920 |
0.36 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4350 |
0.34 |
Binding ≤ 10μM
|
|
CAH13-6-E |
Carbonic Anhydrase XIII (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
9530 |
0.32 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
480 |
0.40 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7400 |
0.33 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8980 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-10-E |
Carbonic Anhydrase VA (cluster #10 Of 10), Eukaryotic |
Eukaryotes |
730 |
0.39 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
890 |
0.39 |
Binding ≤ 10μM
|
|
CAH6-4-E |
Carbonic Anhydrase VI (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9470 |
0.32 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
9820 |
0.32 |
Binding ≤ 10μM
|
|
Z50038-1-O |
Plasmodium Yoelii Yoelii (cluster #1 Of 2), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
470 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
3.38 |
-54.72 |
5 |
4 |
1 |
77 |
302.394 |
7 |
↓
|
|
|
|
Analogs
-
3831448
-
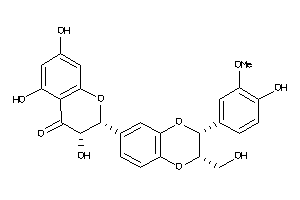
-
4016718
-
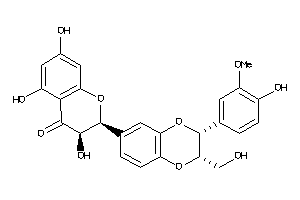
-
5339605
-
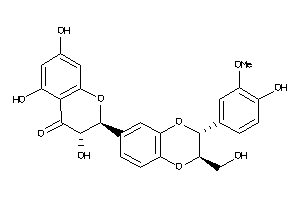
-
8552019
-
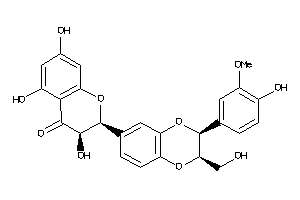
-
14984516
-
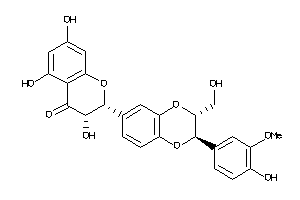
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1490 |
0.23 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9050 |
0.20 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4820 |
0.21 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
2510 |
0.22 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6430 |
0.21 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8960 |
0.20 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4080 |
0.22 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4560 |
0.21 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
9700 |
0.20 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4710 |
0.21 |
Binding ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.21 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.21 |
ADME/T ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
9990 |
0.20 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-8.69 |
-16.42 |
5 |
10 |
0 |
155 |
482.441 |
4 |
↓
|
|
|
|
Analogs
-
4016718
-

-
5339605
-

-
8552019
-

-
17044428
-
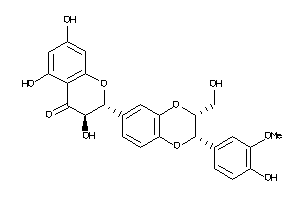
-
17044430
-
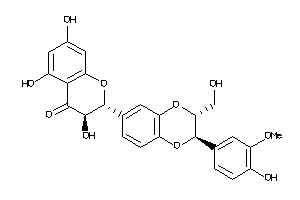
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1490 |
0.23 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9050 |
0.20 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4820 |
0.21 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
2510 |
0.22 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6430 |
0.21 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8960 |
0.20 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4080 |
0.22 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4560 |
0.21 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
9700 |
0.20 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4710 |
0.21 |
Binding ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.21 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.21 |
ADME/T ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
9990 |
0.20 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-8.74 |
-16.51 |
5 |
10 |
0 |
155 |
482.441 |
4 |
↓
|
|
|
|
Analogs
-
4016718
-

-
5339605
-

-
8552019
-

-
17044428
-

-
17044430
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1490 |
0.23 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9050 |
0.20 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4820 |
0.21 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
2510 |
0.22 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6430 |
0.21 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8960 |
0.20 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4080 |
0.22 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4560 |
0.21 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
9700 |
0.20 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4710 |
0.21 |
Binding ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.21 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.21 |
ADME/T ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
9990 |
0.20 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-1.32 |
-16.67 |
5 |
10 |
0 |
155 |
482.441 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.47 |
-0.33 |
-56.57 |
4 |
10 |
-1 |
158 |
481.433 |
4 |
↓
|
|
|
|
Analogs
-
1530850
-
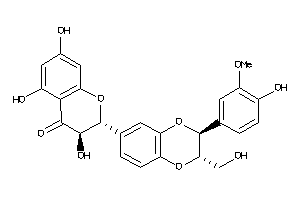
-
1554609
-
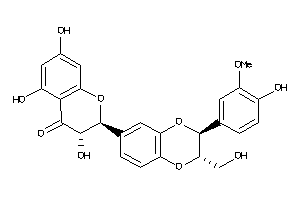
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1490 |
0.23 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9050 |
0.20 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4820 |
0.21 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
650 |
0.25 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
2510 |
0.22 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6430 |
0.21 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
8960 |
0.20 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4080 |
0.22 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4560 |
0.21 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
9700 |
0.20 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4710 |
0.21 |
Binding ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.21 |
Binding ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.21 |
ADME/T ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
9990 |
0.20 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-1.49 |
-26.41 |
5 |
10 |
0 |
155 |
482.441 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.47 |
-0.48 |
-66.96 |
4 |
10 |
-1 |
158 |
481.433 |
4 |
↓
|
|