|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
1.65 |
-13.89 |
0 |
4 |
0 |
39 |
289.338 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.70 |
Binding ≤ 10μM
|
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.65 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.59 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.59 |
Binding ≤ 10μM
|
|
GBRA6-8-E |
GABA Receptor Alpha-6 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
2700 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-8-E |
GABA Receptor Alpha-6 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
2700 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRB2-6-E |
GABA Receptor Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRD-5-E |
GABA Receptor Delta Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRE-5-E |
GABA Receptor Epsilon Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.70 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
6 |
0.64 |
Binding ≤ 10μM
|
|
GBRP-5-E |
GABA Receptor Pi Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
GBRR2-1-E |
GABA Receptor Rho-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
PDE5A-1-E |
Phosphodiesterase 5A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.47 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.63 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
6.49 |
-9.28 |
1 |
4 |
0 |
55 |
240.262 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.04 |
6.94 |
-33.36 |
2 |
4 |
1 |
56 |
241.27 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
7200 |
0.45 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
7200 |
0.45 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
7200 |
0.45 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7200 |
0.45 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7200 |
0.45 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7200 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
5.91 |
-35.08 |
2 |
4 |
0 |
70 |
212.208 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.40 |
5.51 |
-56.26 |
1 |
4 |
-1 |
69 |
211.2 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
|
FABPL-2-E |
Fatty Acid-binding Protein, Liver (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
531 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5600 |
0.37 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.49 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
574 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-4-E |
GABA Receptor Alpha-1 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.58 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
6.91 |
-8.26 |
0 |
3 |
0 |
33 |
284.746 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.74 |
7.78 |
-30.37 |
1 |
3 |
1 |
34 |
285.754 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
245 |
0.42 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
118 |
0.44 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
163 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
-0.67 |
-9.59 |
2 |
4 |
0 |
61 |
296.301 |
4 |
↓
|
|
|
|
Analogs
-
4964600
-
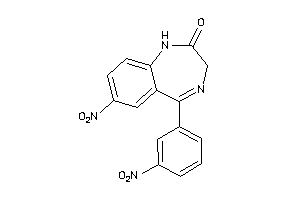
-
5850731
-
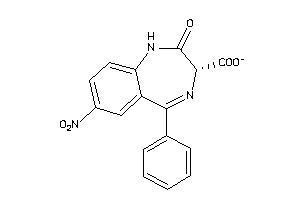
-
4214169
-
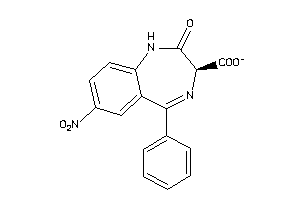
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
38 |
0.49 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
38 |
0.49 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
38 |
0.49 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
38 |
0.49 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
38 |
0.49 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
38 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.14 |
5.47 |
-10.2 |
1 |
6 |
0 |
87 |
281.271 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.14 |
6.34 |
-38.29 |
2 |
6 |
1 |
89 |
282.279 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
660 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
-0.71 |
-10.38 |
2 |
4 |
0 |
61 |
296.301 |
4 |
↓
|
|
|
|
Analogs
-
34929024
-
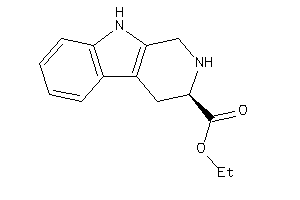
-
793691
-
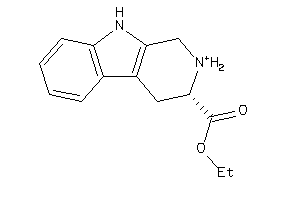
-
4399753
-
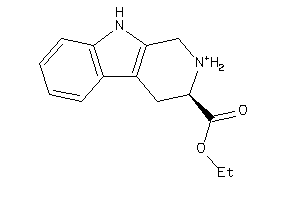
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
5.09 |
-8.09 |
2 |
4 |
0 |
54 |
244.294 |
3 |
↓
|
|
|
|
Analogs
-
34929023
-
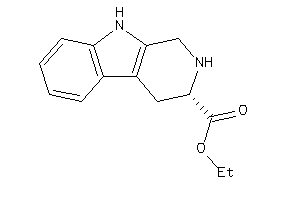
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
5.11 |
-7.45 |
2 |
4 |
0 |
54 |
244.294 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
29 |
0.53 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
29 |
0.53 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
29 |
0.53 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
29 |
0.53 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
29 |
0.53 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
29 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
8.07 |
-8.93 |
1 |
4 |
0 |
55 |
268.316 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.10 |
8.52 |
-33.46 |
2 |
4 |
1 |
56 |
269.324 |
5 |
↓
|
|
|
|
Analogs
-
6582775
-
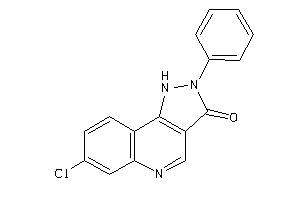
-
13448368
-
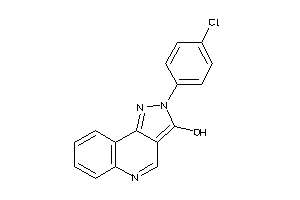
-
20744554
-
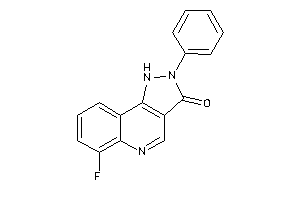
-
26507437
-
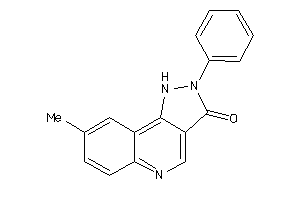
-
26507550
-
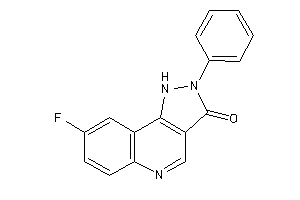
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
200 |
0.47 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
17 |
0.54 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.54 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
46 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
7.11 |
-8.83 |
0 |
4 |
0 |
45 |
261.284 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
2.85 |
6.99 |
-12.47 |
1 |
4 |
0 |
51 |
261.284 |
1 |
↓
|
|
|
|
Analogs
-
34925907
-
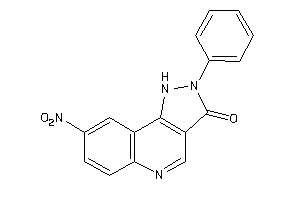
-
6116830
-
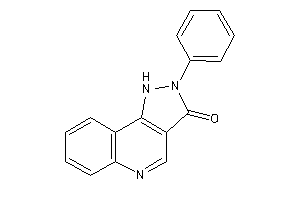
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
7.63 |
-7.18 |
0 |
4 |
0 |
45 |
295.729 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
3.52 |
7.51 |
-11.39 |
1 |
4 |
0 |
51 |
295.729 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
285 |
0.46 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
285 |
0.46 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
285 |
0.46 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
285 |
0.46 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
285 |
0.46 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
285 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
5.61 |
-8.91 |
2 |
4 |
0 |
62 |
284.34 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
105 |
0.47 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
105 |
0.47 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
105 |
0.47 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
105 |
0.47 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
105 |
0.47 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
105 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
4.71 |
-10.65 |
2 |
5 |
0 |
75 |
279.299 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
324 |
0.45 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
324 |
0.45 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
324 |
0.45 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
324 |
0.45 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
324 |
0.45 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
324 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
4.62 |
-9.89 |
2 |
5 |
0 |
75 |
268.272 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.68 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.67 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.68 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
58 |
0.60 |
Binding ≤ 10μM
|
|
GBRA6-8-E |
GABA Receptor Alpha-6 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.67 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRB2-6-E |
GABA Receptor Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.67 |
Binding ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRD-5-E |
GABA Receptor Delta Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRE-5-E |
GABA Receptor Epsilon Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.70 |
Binding ≤ 10μM
|
|
GBRP-5-E |
GABA Receptor Pi Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
GBRR2-1-E |
GABA Receptor Rho-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.69 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.68 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
5.56 |
-9.44 |
1 |
4 |
0 |
55 |
226.235 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.66 |
6.01 |
-33.94 |
2 |
4 |
1 |
56 |
227.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
7.29 |
-9.04 |
1 |
4 |
0 |
55 |
254.289 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.54 |
7.74 |
-33.43 |
2 |
4 |
1 |
56 |
255.297 |
4 |
↓
|
|
|
|
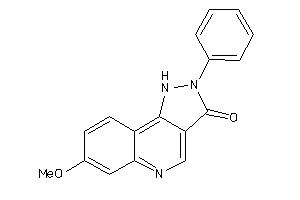
Analogs
-
13741323
-
-
35786837
-
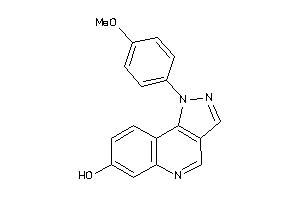
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
4.54 |
-12.43 |
1 |
5 |
0 |
60 |
291.31 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.90 |
6.28 |
-12.66 |
1 |
5 |
0 |
60 |
291.31 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
195 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1950 |
0.35 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
163 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
163 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
163 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
163 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
-1.39 |
-10.17 |
2 |
5 |
0 |
71 |
308.337 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.39 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.39 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.39 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
94 |
0.39 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
94 |
0.39 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
94 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
5.69 |
-12.54 |
2 |
6 |
0 |
80 |
338.363 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
8.7 |
-9.48 |
1 |
4 |
0 |
51 |
326.783 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
2600 |
0.36 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
2600 |
0.36 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
2600 |
0.36 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2600 |
0.36 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2600 |
0.36 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2600 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
6.91 |
-9.6 |
2 |
4 |
0 |
62 |
312.756 |
4 |
↓
|
|
|
|
Analogs
-
3631205
-
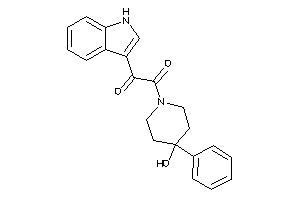
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1260 |
0.36 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1260 |
0.36 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
1260 |
0.36 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1260 |
0.36 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1260 |
0.36 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1260 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
8.05 |
-9.65 |
2 |
4 |
0 |
62 |
306.365 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
-1.39 |
-10.3 |
2 |
5 |
0 |
71 |
308.337 |
5 |
↓
|
|
|
|
Analogs
-
35076415
-
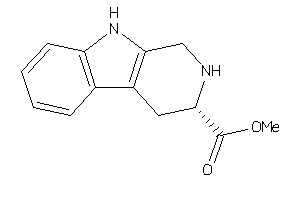
-
193844
-
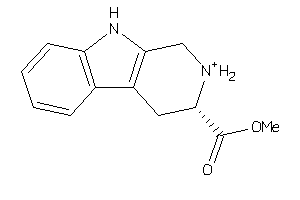
-
193842
-
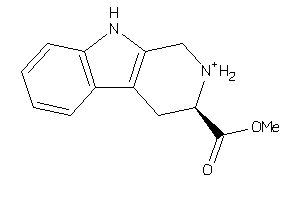
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
4.18 |
-7.67 |
2 |
4 |
0 |
54 |
230.267 |
2 |
↓
|
|
|
|
Analogs
-
193844
-

-
34924544
-
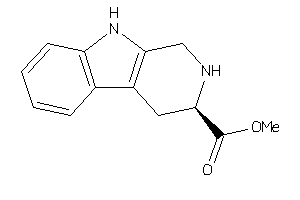
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
250 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
4.17 |
-8.3 |
2 |
4 |
0 |
54 |
230.267 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
-2.38 |
-11.04 |
2 |
5 |
0 |
74 |
279.299 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.94 |
-2.32 |
-38.4 |
3 |
5 |
1 |
76 |
280.307 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
426 |
0.42 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
426 |
0.42 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
426 |
0.42 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
426 |
0.42 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
426 |
0.42 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
426 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
-2.18 |
-10.54 |
2 |
5 |
0 |
74 |
279.299 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
106 |
0.44 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.55 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
148 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
94 |
0.45 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
148 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
58 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
8.07 |
-13.1 |
0 |
6 |
0 |
64 |
303.293 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2700 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.83 |
7.37 |
-13.06 |
1 |
6 |
0 |
70 |
352.39 |
6 |
↓
|
|
|
|
Analogs
-
6096054
-
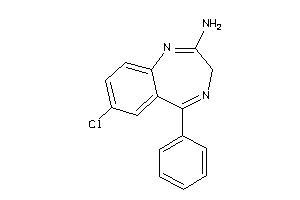
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
770 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
460 |
0.42 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
740 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
520 |
0.42 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
770 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
438 |
0.42 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
910 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
7.54 |
-13.92 |
1 |
4 |
0 |
53 |
299.761 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
7.84 |
-45.23 |
2 |
4 |
1 |
55 |
300.769 |
1 |
↓
|
|
|
|
Analogs
-
4214169
-

-
5850731
-

-
6095266
-
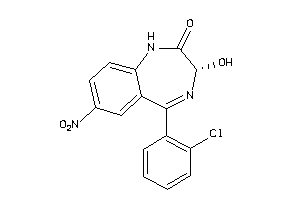
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
2 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
-2.23 |
-10.75 |
1 |
6 |
0 |
87 |
315.716 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-2-E |
GABA Receptor Alpha-1 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
|
GBRA2-2-E |
GABA Receptor Alpha-2 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-2-E |
GABA Receptor Alpha-3 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-2-E |
GABA Receptor Alpha-4 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
3.23 |
-10.97 |
0 |
2 |
0 |
34 |
282.383 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
90 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
6.57 |
-11.14 |
2 |
5 |
0 |
71 |
322.364 |
6 |
↓
|
|
|
|
Analogs
-
4163077
-
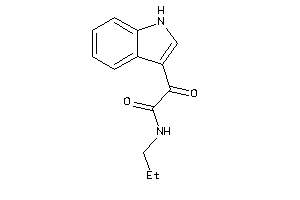
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
7.27 |
-9.42 |
2 |
4 |
0 |
62 |
292.338 |
5 |
↓
|
|
|
|
Analogs
-
4163077
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1500 |
0.33 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1500 |
0.33 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
1500 |
0.33 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1500 |
0.33 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1500 |
0.33 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1500 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
7.33 |
-12.35 |
3 |
5 |
0 |
78 |
331.375 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
340 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
7.79 |
-9.95 |
2 |
4 |
0 |
62 |
326.783 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
85 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
6.57 |
-11.12 |
2 |
5 |
0 |
71 |
322.364 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
700 |
0.32 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
700 |
0.32 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
700 |
0.32 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
700 |
0.32 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
700 |
0.32 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
700 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
6.99 |
-13.97 |
2 |
6 |
0 |
80 |
386.835 |
7 |
↓
|
|
|
|
Analogs
-
6458196
-
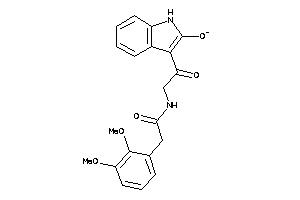
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
510 |
0.34 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
510 |
0.34 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
510 |
0.34 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
510 |
0.34 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.34 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
510 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
6.47 |
-13.65 |
2 |
6 |
0 |
80 |
352.39 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
67 |
0.46 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
67 |
0.46 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
67 |
0.46 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
67 |
0.46 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
67 |
0.46 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
67 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
7.02 |
-8.97 |
2 |
4 |
0 |
62 |
312.756 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
107 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
107 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
107 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
107 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
107 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
107 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
6.32 |
-10.61 |
2 |
5 |
0 |
71 |
342.782 |
5 |
↓
|
|
|
|
Analogs
-
26380302
-
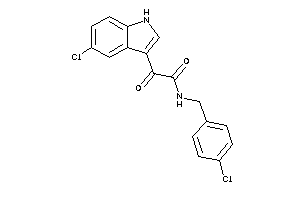
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
490 |
0.40 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
490 |
0.40 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
490 |
0.40 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
490 |
0.40 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
490 |
0.40 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
490 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
7.02 |
-9.17 |
2 |
4 |
0 |
62 |
312.756 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
100 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
7.86 |
-9.18 |
2 |
4 |
0 |
62 |
326.783 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
30 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
6.21 |
-13.04 |
2 |
6 |
0 |
80 |
372.808 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
346 |
0.43 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
120 |
0.46 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
120 |
0.46 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
120 |
0.46 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
120 |
0.46 |
Binding ≤ 10μM
|
|
GBRB2-6-E |
GABA Receptor Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
346 |
0.43 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
120 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
-1.41 |
-8.78 |
2 |
4 |
0 |
61 |
278.311 |
4 |
↓
|
|
|
|
Analogs
-
4163159
-
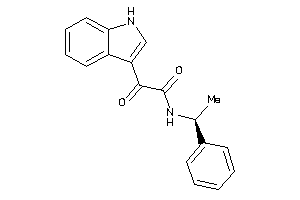
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1150 |
0.38 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
1550 |
0.37 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
1307 |
0.37 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1307 |
0.37 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1307 |
0.37 |
Binding ≤ 10μM
|
|
GBRB2-6-E |
GABA Receptor Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
1150 |
0.38 |
Binding ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5500 |
0.33 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1307 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
-0.65 |
-8.81 |
2 |
4 |
0 |
61 |
292.338 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
494 |
0.35 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
494 |
0.35 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
494 |
0.35 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
494 |
0.35 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
494 |
0.35 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
494 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
5.09 |
-13.12 |
2 |
6 |
0 |
80 |
338.363 |
6 |
↓
|
|