|
|
Analogs
-
120294
-
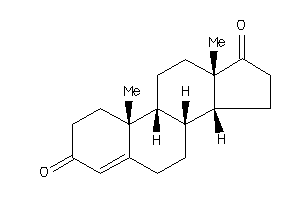
-
2046798
-
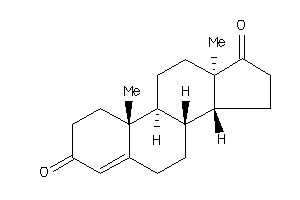
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Binding ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ESR2-2-E |
Estrogen Receptor Beta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.46 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
31 |
0.46 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.48 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.34 |
Binding ≤ 10μM
|
|
SGMR1-2-E |
Sigma Opioid Receptor (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
260 |
0.40 |
Binding ≤ 10μM
|
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
37 |
0.45 |
Functional ≤ 10μM
|
|
ESR1-2-E |
Estrogen Receptor Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.30 |
Functional ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2770 |
0.34 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Functional ≤ 10μM
|
|
PRGR-2-E |
Progesterone Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Functional ≤ 10μM
|
|
CP2C9-2-E |
Cytochrome P450 2C9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
ADME/T ≤ 10μM
|
|
ERG2-1-F |
C-8 Sterol Isomerase (cluster #1 Of 2), Fungal |
Fungi |
4430 |
0.33 |
Binding ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
6310 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-1-O |
T47D (Breast Carcinoma Cells) (cluster #1 Of 7), Other |
Other |
1 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
10.64 |
-9.9 |
0 |
2 |
0 |
34 |
314.469 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-1-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
580 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.02 |
-1.02 |
-56.39 |
2 |
3 |
1 |
39 |
395.935 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.02 |
-0.94 |
-110.28 |
3 |
3 |
2 |
40 |
396.943 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
12.27 |
-52.74 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.73 |
10.3 |
-5.37 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
-
3978828
-
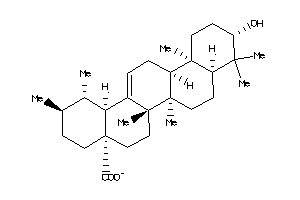
-
3978829
-
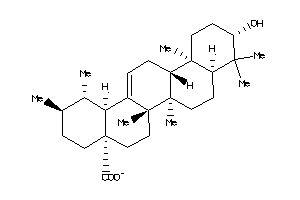
-
4273370
-
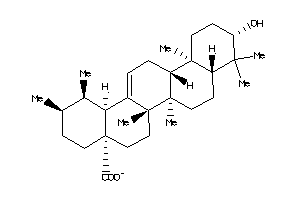
-
4273371
-
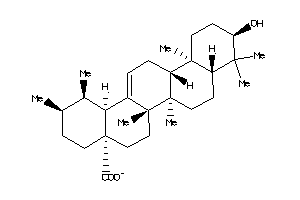
-
4273372
-
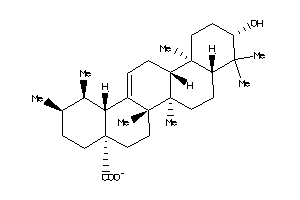
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.22 |
Binding ≤ 10μM
|
|
PA21B-3-E |
Phospholipase A2 Group 1B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.23 |
Binding ≤ 10μM
|
|
PA2A-1-E |
Phospholipase A2 Isozyme PLA-A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GA-3-E |
Phospholipase A2, Membrane Associated (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GD-2-E |
Group IID Secretory Phospholipase A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GE-2-E |
Group IIE Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GF-2-E |
Group IIF Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-2-E |
T-cell Protein-tyrosine Phosphatase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6700 |
0.22 |
Binding ≤ 10μM |
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.21 |
Binding ≤ 10μM |
|
Q7T3S7-1-E |
Phospholipase A2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.25 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-4-O |
Trypanosoma Brucei (cluster #4 Of 6), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50420-3-O |
Trypanosoma Brucei Brucei (cluster #3 Of 7), Other |
Other |
2200 |
0.24 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50472-2-O |
Toxoplasma Gondii (cluster #2 Of 4), Other |
Other |
1000 |
0.25 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
1800 |
0.24 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
12.05 |
-49.78 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
10.29 |
-5.45 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
3.58 |
-52.11 |
3 |
5 |
-1 |
101 |
407.571 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3914812
-
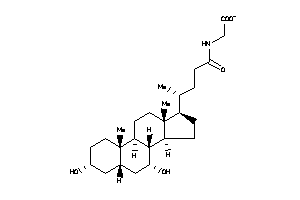
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1180 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
-4.68 |
-47.75 |
3 |
6 |
-1 |
109 |
448.624 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2620 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.33 |
-5.82 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.23 |
8.02 |
-48.98 |
2 |
4 |
-1 |
81 |
419.626 |
5 |
↓
|
|
|
|
Analogs
-
4716552
-
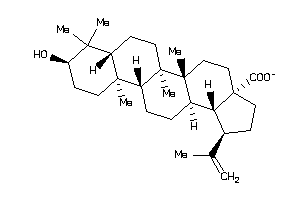
-
4995154
-
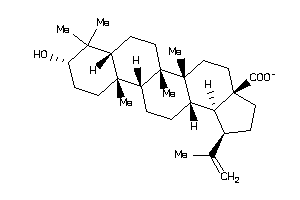
-
4995155
-
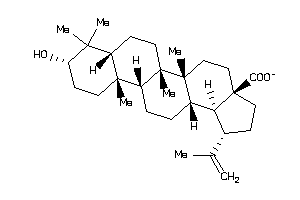
-
4995156
-
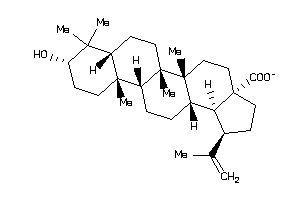
-
8951991
-
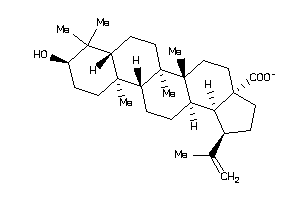
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.22 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2170 |
0.24 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z103202-1-O |
8505C (cluster #1 Of 2), Other |
Other |
7260 |
0.22 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
5200 |
0.22 |
Functional ≤ 10μM
|
|
Z50459-6-O |
Leishmania Donovani (cluster #6 Of 8), Other |
Other |
4100 |
0.23 |
Functional ≤ 10μM
|
|
Z50602-3-O |
Human Herpesvirus 1 (cluster #3 Of 5), Other |
Other |
8200 |
0.22 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
3100 |
0.23 |
Functional ≤ 10μM
|
|
Z50636-1-O |
Sindbis Virus (cluster #1 Of 3), Other |
Other |
500 |
0.27 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
7000 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
6480 |
0.22 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
6650 |
0.22 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
1400 |
0.25 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
900 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.04 |
12.1 |
-48.88 |
1 |
3 |
-1 |
60 |
455.703 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.04 |
10.34 |
-5.48 |
2 |
3 |
0 |
58 |
456.711 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.36 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
4.35 |
-8.21 |
3 |
3 |
0 |
61 |
378.597 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
G6PD-1-E |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.36 |
Binding ≤ 10μM
|
|
Q9GRG7-1-E |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
490 |
0.42 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3200 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.76 |
-6.06 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4390 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
-0.4 |
-8.32 |
2 |
4 |
0 |
66 |
406.607 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
8762630
-
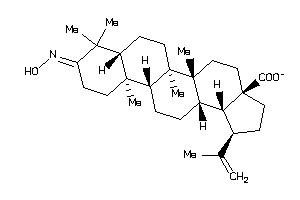
-
8762634
-
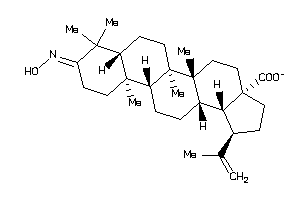
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.31 |
11.6 |
-54 |
1 |
4 |
-1 |
73 |
468.702 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
3831599
-
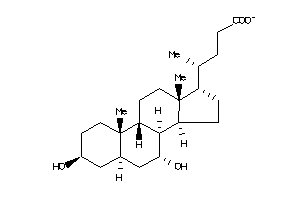
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1250 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
7.2 |
-49.51 |
2 |
4 |
-1 |
81 |
391.572 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.25 |
5.22 |
-9.71 |
3 |
4 |
0 |
78 |
392.58 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
270 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.98 |
0.51 |
-55.63 |
0 |
3 |
-1 |
57 |
373.557 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
5751225
-
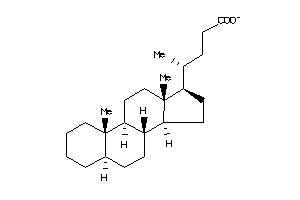
-
4577159
-
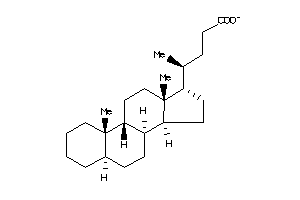
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6080 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.55 |
2.61 |
-45.67 |
0 |
2 |
-1 |
40 |
359.574 |
4 |
↓
|
|
|
|
Analogs
-
8762249
-
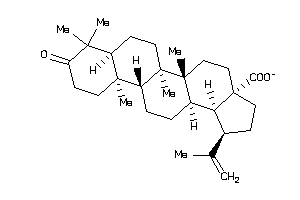
-
8762252
-
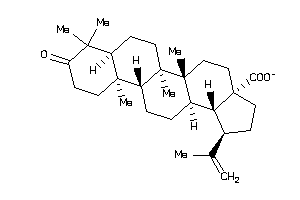
-
8835967
-
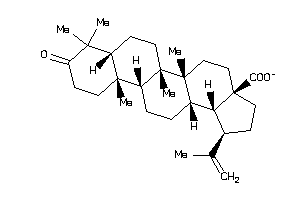
-
8835968
-
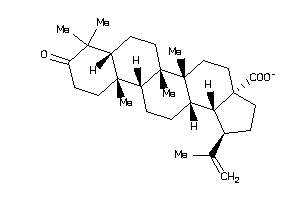
-
8952537
-
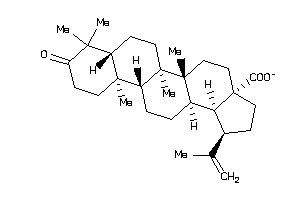
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4710 |
0.23 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z100499-2-O |
SARS Coronavirus (cluster #2 Of 2), Other |
Other |
630 |
0.26 |
Functional ≤ 10μM
|
|
Z103205-2-O |
A431 (cluster #2 Of 4), Other |
Other |
3260 |
0.23 |
Functional ≤ 10μM
|
|
Z50602-3-O |
Human Herpesvirus 1 (cluster #3 Of 5), Other |
Other |
2500 |
0.24 |
Functional ≤ 10μM
|
|
Z50652-2-O |
Influenza A Virus (cluster #2 Of 4), Other |
Other |
5700 |
0.22 |
Functional ≤ 10μM
|
|
Z80186-4-O |
K562 (Erythroleukemia Cells) (cluster #4 Of 11), Other |
Other |
6000 |
0.22 |
Functional ≤ 10μM
|
|
Z80928-4-O |
HCT-116 (Colon Carcinoma Cells) (cluster #4 Of 9), Other |
Other |
2610 |
0.24 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
2980 |
0.23 |
Functional ≤ 10μM
|
|
Z81252-4-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #4 Of 11), Other |
Other |
9770 |
0.21 |
Functional ≤ 10μM
|
|
Z80186-2-O |
K562 (Erythroleukemia Cells) (cluster #2 Of 3), Other |
Other |
6000 |
0.22 |
ADME/T ≤ 10μM
|
|
Z81115-3-O |
KB (Squamous Cell Carcinoma) (cluster #3 Of 3), Other |
Other |
3800 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.86 |
14.43 |
-48.14 |
0 |
3 |
-1 |
57 |
453.687 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.86 |
12.66 |
-7.5 |
1 |
3 |
0 |
54 |
454.695 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5aR,5bR,7aR,9R,11aR,11bR,13aR,13bR)-9-allyl-9-hydroxy-1-isopropenyl-5a,5b,8,8,11a-pentamethy
(1R,3aS,5aR,5bR,7aR,9R,11aR,11bR…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.27 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.03 |
13.9 |
-48.33 |
1 |
3 |
-1 |
60 |
495.768 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.03 |
12.16 |
-5.05 |
2 |
3 |
0 |
58 |
496.776 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB3-3-E |
Estradiol 17-beta-dehydrogenase 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
330 |
0.43 |
Binding ≤ 10μM
|
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4140 |
0.36 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3760 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.62 |
-6.45 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-2-E |
GABA Receptor Alpha-5 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-2-E |
GABA Receptor Beta-1 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRG3-3-E |
GABA Receptor Gamma-3 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
Q91ZM7-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
410 |
0.43 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6220 |
0.35 |
Functional ≤ 10μM
|
|
Z104301-2-O |
GABA-A Receptor; Anion Channel (cluster #2 Of 8), Other |
Other |
1367 |
0.39 |
Binding ≤ 10μM
|
|
Z50573-2-O |
Xenopus Laevis (cluster #2 Of 2), Other |
Other |
3380 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
6.9 |
-6.34 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
|