|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
106 |
0.41 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
77 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
7.01 |
-12.95 |
0 |
4 |
0 |
48 |
325.408 |
3 |
↓
|
|
|
|
Analogs
-
13676104
-
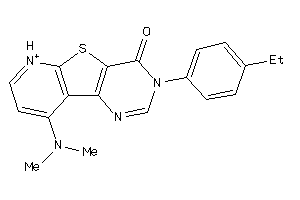
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
890 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
10.48 |
-39.44 |
1 |
5 |
1 |
52 |
337.428 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.97 |
10.05 |
-14.15 |
0 |
5 |
0 |
51 |
336.42 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.47 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
20 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
9.79 |
-39.6 |
1 |
5 |
1 |
52 |
323.401 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1030 |
0.35 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
16 |
0.45 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1810 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
-1.46 |
-41.13 |
1 |
5 |
1 |
52 |
357.846 |
2 |
↓
|
|
|
|
Analogs
-
22066252
-
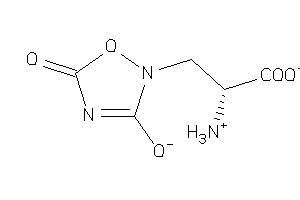
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FOLH1-1-E |
Glutamate Carboxypeptidase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.54 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA2-4-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA3-4-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIA4-4-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.76 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
171 |
0.73 |
Binding ≤ 10μM
|
|
GRIK2-3-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1670 |
0.62 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRIK5-1-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
200 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
150 |
0.73 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
GRM8-2-E |
Metabotropic Glutamate Receptor 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T35-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T43-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
Q80T47-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Binding ≤ 10μM
|
|
XCT-1-E |
Cystine/glutamate Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.57 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.64 |
Functional ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
55 |
0.78 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
7300 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.04 |
-4.9 |
-43.55 |
3 |
8 |
-1 |
139 |
188.119 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.04 |
-5.21 |
-87.71 |
2 |
8 |
-2 |
137 |
187.111 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.04 |
-7.81 |
-15.85 |
4 |
8 |
0 |
132 |
189.127 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIK1-1-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2430 |
0.71 |
Binding ≤ 10μM |
|
GRIK2-3-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3230 |
0.70 |
Binding ≤ 10μM |
|
GRIK3-3-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1740 |
0.73 |
Binding ≤ 10μM |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.66 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8000 |
0.65 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.66 |
Functional ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.66 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
10000 |
0.64 |
Binding ≤ 10μM
|
|
Z50597-8-O |
Rattus Norvegicus (cluster #8 Of 12), Other |
Other |
9600 |
0.64 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.16 |
-1.99 |
-41.18 |
3 |
6 |
-1 |
112 |
157.105 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.70 |
-3.67 |
-101.23 |
2 |
6 |
-2 |
115 |
156.097 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.70 |
-4.09 |
-41.28 |
4 |
6 |
0 |
114 |
158.113 |
2 |
↓
|
|
|
|
Analogs
-
184798
-
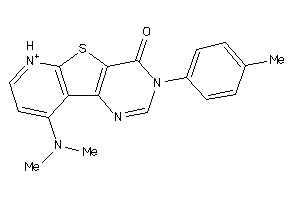
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
194 |
0.38 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
442 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.72 |
-40.51 |
1 |
5 |
1 |
52 |
351.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
342 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
6.77 |
-40.44 |
1 |
6 |
1 |
56 |
344.464 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.41 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
281 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
10.38 |
-41.41 |
1 |
5 |
1 |
52 |
337.428 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.85 |
9.94 |
-17.36 |
0 |
5 |
0 |
51 |
336.42 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.45 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.45 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
9.09 |
-40.94 |
1 |
6 |
1 |
62 |
353.427 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.89 |
12.17 |
-19.07 |
1 |
5 |
0 |
60 |
368.44 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8990 |
0.27 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.36 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
GRM4-2-E |
Metabotropic Glutamate Receptor 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2650 |
0.30 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1140 |
0.32 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.28 |
Functional ≤ 10μM
|
|
GRM2-2-E |
Metabotropic Glutamate Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.41 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.42 |
Functional ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8200 |
0.27 |
Functional ≤ 10μM
|
|
GRM7-2-E |
Metabotropic Glutamate Receptor 7 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
990 |
0.32 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
170 |
0.36 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
10 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
10.29 |
-80.49 |
3 |
6 |
-1 |
117 |
352.366 |
5 |
↓
|
|
|
|
Analogs
-
4350130
-
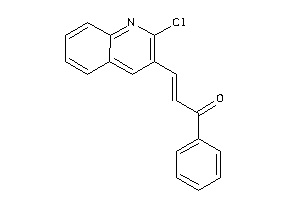
-
5799012
-
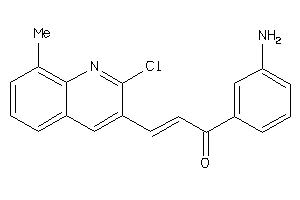
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9180 |
0.32 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1700 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
1.43 |
-12.93 |
0 |
2 |
0 |
29 |
307.78 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9930 |
0.33 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
740 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
0.61 |
-11.39 |
0 |
2 |
0 |
29 |
313.809 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 145 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
613 |
0.87 |
Binding ≤ 10μM
|
|
EAA3-2-E |
Excitatory Amino Acid Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
51 |
1.02 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1360 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.87 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.77 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.79 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
4100 |
0.75 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.72 |
Binding ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.77 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
450 |
0.89 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6160 |
0.73 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.73 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3660 |
0.76 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.70 |
Functional ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.71 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.71 |
Functional ≤ 10μM
|
|
GRM4-1-E |
Metabotropic Glutamate Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.73 |
Functional ≤ 10μM
|
|
GRM5-5-E |
Metabotropic Glutamate Receptor 5 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7600 |
0.72 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.70 |
Functional ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.80 |
Functional ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.80 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
870 |
0.85 |
Binding ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 5), Other |
Other |
170 |
0.95 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
1300 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.74 |
-73.91 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 120 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MURI-2-B |
Glutamate Racemase (cluster #2 Of 2), Bacterial |
Bacteria |
5800 |
0.73 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1362 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.92 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.80 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9800 |
0.70 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3100 |
0.77 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1050 |
0.84 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
80 |
0.99 |
Binding ≤ 10μM |
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
2500 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.72 |
-74 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
736 |
0.45 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
713 |
0.45 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
96 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
9.46 |
-5.82 |
1 |
3 |
0 |
38 |
318.149 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
116 |
0.51 |
Binding ≤ 10μM
|
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
398 |
0.47 |
Functional ≤ 10μM
|
|
GRM5-1-E |
Metabotropic Glutamate Receptor 5 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
256 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
9.9 |
-5.54 |
1 |
3 |
0 |
38 |
334.604 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
104 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
-2.69 |
-10.95 |
2 |
4 |
0 |
54 |
292.338 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
69 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
5.88 |
-9.81 |
1 |
4 |
0 |
55 |
277.327 |
4 |
↓
|
|
|
|
Analogs
-
4881873
-
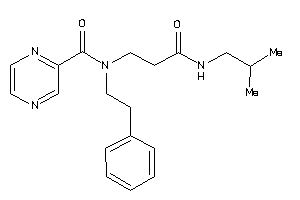
-
12931374
-
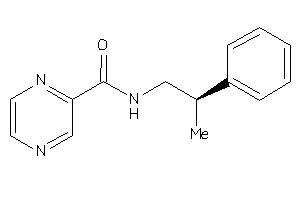
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
6550 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
4.92 |
-9.73 |
1 |
4 |
0 |
55 |
255.321 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
5.67 |
-7.6 |
1 |
5 |
0 |
58 |
302.422 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
0.21 |
-7.99 |
0 |
4 |
0 |
52 |
292.386 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7000 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
4.34 |
-7.73 |
1 |
4 |
0 |
55 |
257.337 |
2 |
↓
|
|
|
|
Analogs
-
545889
-
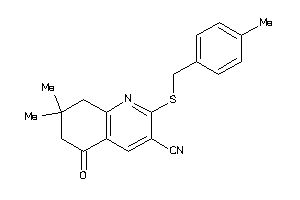
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4800 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.32 |
2.2 |
-8.86 |
0 |
3 |
0 |
53 |
322.433 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.66 |
2.6 |
-75.73 |
3 |
5 |
-1 |
108 |
172.16 |
2 |
↓
|
|
|
|
Analogs
-
2379561
-
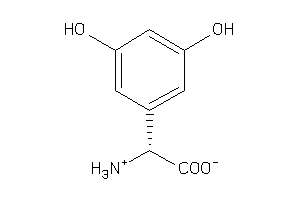
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.65 |
Binding ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6600 |
0.56 |
Functional ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1900 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.36 |
-2.45 |
-39.02 |
5 |
5 |
0 |
108 |
183.163 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
-2.96 |
-8.13 |
1 |
4 |
0 |
54 |
307.397 |
2 |
↓
|
|
|
|
Analogs
-
36880408
-
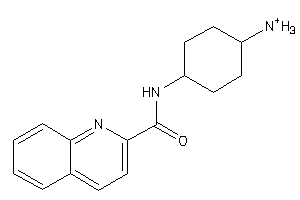
-
36880540
-
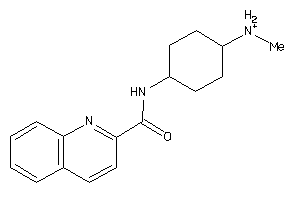
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-2-E |
Metabotropic Glutamate Receptor 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
440 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.13 |
-1.88 |
-7.7 |
1 |
3 |
0 |
41 |
306.409 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
37984152
-
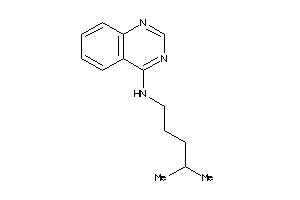
-
42187696
-
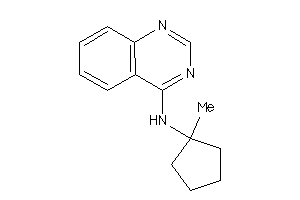
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
8.2 |
-8.89 |
1 |
3 |
0 |
38 |
227.311 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.42 |
8.65 |
-29.1 |
2 |
3 |
1 |
39 |
228.319 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2970 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
0.61 |
-12.02 |
0 |
2 |
0 |
29 |
313.809 |
3 |
↓
|
|
|
|
Analogs
-
5799012
-

-
38418430
-
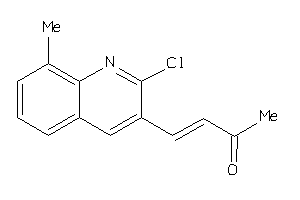
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8500 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.99 |
1.6 |
-13.33 |
0 |
2 |
0 |
29 |
307.78 |
3 |
↓
|
|