|
|
Analogs
-
13455558
-
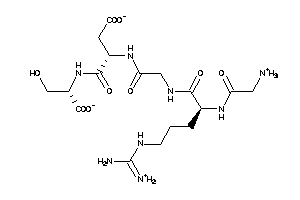
-
33716798
-
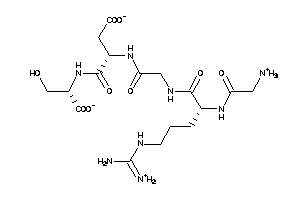
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.23 |
Binding ≤ 10μM |
|
ITA5-2-E |
Fibronectin Receptor Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
|
ITB1-2-E |
Fibronectin Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.26 |
Binding ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9600 |
0.23 |
Binding ≤ 10μM |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.25 |
Functional ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.37 |
-4.78 |
-154.71 |
12 |
15 |
0 |
279 |
433.422 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.37 |
-5.12 |
-121.45 |
11 |
15 |
-1 |
277 |
432.414 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-5.37 |
-6.79 |
-127.6 |
13 |
15 |
1 |
276 |
434.43 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.14 |
-1.21 |
-147.1 |
11 |
14 |
0 |
261 |
449.49 |
14 |
↓
|
|
Ref
Reference (pH 7)
|
-5.10 |
-1.2 |
-146.16 |
11 |
14 |
0 |
259 |
449.49 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-5.14 |
-3.42 |
-106.32 |
10 |
14 |
-1 |
260 |
448.482 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
76 |
0.42 |
Binding ≤ 10μM |
|
ITA5-2-E |
Fibronectin Receptor Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
510 |
0.37 |
Binding ≤ 10μM |
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
76 |
0.42 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.93 |
-2.53 |
-153.33 |
10 |
12 |
0 |
232 |
346.344 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM |
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.24 |
Binding ≤ 10μM |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Functional ≤ 10μM |
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Functional ≤ 10μM |
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 1), Other |
Other |
6000 |
0.24 |
Binding ≤ 10μM |
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.79 |
0.21 |
-176.06 |
11 |
14 |
0 |
259 |
445.477 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.79 |
0.11 |
-139.7 |
10 |
14 |
-1 |
257 |
444.469 |
15 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-4.79 |
-1.54 |
-133.75 |
12 |
14 |
1 |
256 |
446.485 |
15 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.86 |
4.16 |
-46.86 |
5 |
10 |
1 |
143 |
421.474 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
-0.99 |
4.3 |
-19.23 |
4 |
10 |
0 |
144 |
420.466 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.86 |
5.25 |
-32.88 |
4 |
10 |
0 |
146 |
420.466 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.16 |
Binding ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.16 |
Binding ≤ 10μM
|
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
210 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.75 |
-2.85 |
-194.48 |
17 |
22 |
1 |
385 |
716.774 |
24 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.77 |
-3.21 |
-154.73 |
16 |
22 |
0 |
386 |
715.766 |
23 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.75 |
-3.25 |
-155.07 |
16 |
22 |
0 |
384 |
715.766 |
24 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
-5.37 |
-56.05 |
6 |
9 |
1 |
139 |
362.41 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
-1.97 |
-43.86 |
6 |
8 |
1 |
136 |
359.406 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
8.66 |
-69.9 |
3 |
7 |
0 |
112 |
440.606 |
14 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
6.41 |
-48.25 |
4 |
8 |
0 |
131 |
442.475 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
-3.5 |
-72.59 |
1 |
6 |
0 |
69 |
355.438 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
6.59 |
-56.84 |
0 |
4 |
-1 |
66 |
179.151 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-0.93 |
3.82 |
-67.39 |
4 |
7 |
0 |
101 |
345.447 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.93 |
5.98 |
-99.31 |
5 |
7 |
1 |
103 |
346.455 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
-5.89 |
-85.8 |
3 |
7 |
0 |
112 |
440.606 |
14 |
↓
|
|