|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.32 |
Functional ≤ 10μM |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.97 |
-19.98 |
0 |
8 |
0 |
80 |
450.93 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
395 |
0.30 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
395 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
12.5 |
-10.63 |
0 |
4 |
0 |
39 |
401.506 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
395 |
0.30 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
395 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
12.6 |
-11.99 |
0 |
4 |
0 |
39 |
401.506 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
900 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
10.7 |
-12.83 |
1 |
6 |
0 |
68 |
444.531 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
10.53 |
-14.56 |
1 |
6 |
0 |
68 |
444.531 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.36 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
7.7 |
-13.37 |
1 |
6 |
0 |
64 |
397.519 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.26 |
8.16 |
-41.21 |
2 |
6 |
1 |
65 |
398.527 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.45 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3162 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.19 |
-3.47 |
-5.9 |
2 |
5 |
0 |
59 |
484.188 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.35 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.71 |
14.58 |
-20.32 |
1 |
6 |
0 |
67 |
470.598 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.71 |
15 |
-40.24 |
2 |
6 |
1 |
68 |
471.606 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.32 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
14.82 |
-22.92 |
1 |
6 |
0 |
67 |
452.558 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.34 |
15.21 |
-37.76 |
2 |
6 |
1 |
68 |
453.566 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.30 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
12.44 |
-12.26 |
1 |
5 |
0 |
51 |
512.572 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.45 |
12.54 |
-47.4 |
2 |
5 |
1 |
52 |
513.58 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
346 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
8.96 |
-11.55 |
0 |
5 |
0 |
58 |
378.469 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.37 |
Binding ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
8.1 |
-15.49 |
0 |
7 |
0 |
76 |
420.531 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
232 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
9.57 |
-12.96 |
0 |
5 |
0 |
58 |
374.506 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
661 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
8.19 |
-13.24 |
0 |
6 |
0 |
67 |
390.505 |
8 |
↓
|
|
|
|
Analogs
-
4860424
-
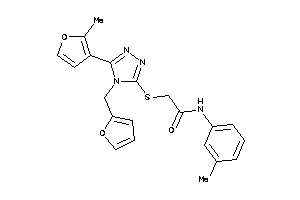
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
794 |
0.28 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.41 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.27 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.69 |
-15.25 |
1 |
7 |
0 |
86 |
422.51 |
7 |
↓
|
|
|
|
Analogs
-
4860424
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
631 |
0.29 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.64 |
-14.93 |
1 |
7 |
0 |
86 |
422.51 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1259 |
0.28 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.92 |
-15.07 |
1 |
7 |
0 |
86 |
422.51 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
398 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.97 |
-14.83 |
1 |
7 |
0 |
86 |
422.51 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2512 |
0.25 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
16 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
10.52 |
-14.99 |
1 |
8 |
0 |
95 |
438.509 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.28 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.09 |
10.99 |
-16.94 |
1 |
7 |
0 |
86 |
446.891 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
2.01 |
-14.91 |
1 |
7 |
0 |
86 |
394.456 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
2.27 |
-15.17 |
1 |
7 |
0 |
86 |
408.483 |
7 |
↓
|
|
|
|
Analogs
-
13512057
-
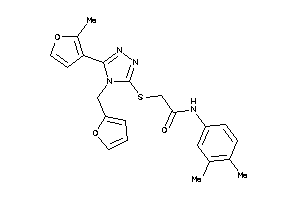
-
13512099
-
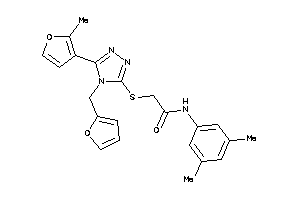
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2512 |
0.27 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
2.27 |
-15.17 |
1 |
7 |
0 |
86 |
408.483 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
158 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
2.75 |
-15.21 |
1 |
7 |
0 |
86 |
412.446 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
2.69 |
-17.25 |
1 |
7 |
0 |
86 |
412.446 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1995 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
2.8 |
-14.07 |
1 |
7 |
0 |
86 |
412.446 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1995 |
0.28 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.00 |
10.94 |
-14.65 |
1 |
7 |
0 |
86 |
428.901 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1995 |
0.28 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
63 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
10.87 |
-16.28 |
1 |
7 |
0 |
86 |
428.901 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
9.85 |
-14.88 |
1 |
8 |
0 |
95 |
424.482 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
9.64 |
-16.55 |
1 |
8 |
0 |
95 |
424.482 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2512 |
0.25 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
10.83 |
-20.71 |
1 |
8 |
0 |
103 |
436.493 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3162 |
0.25 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
316 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
10.75 |
-23.31 |
1 |
8 |
0 |
103 |
436.493 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
794 |
0.28 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
158 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
10.75 |
-16.14 |
1 |
8 |
0 |
89 |
437.525 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1585 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.42 |
11.32 |
-15.47 |
1 |
7 |
0 |
86 |
422.51 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.29 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
17 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
10.38 |
-22.97 |
2 |
8 |
0 |
100 |
535.695 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.35 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.78 |
-30.94 |
3 |
8 |
0 |
106 |
487.01 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.34 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
10.69 |
-31.38 |
2 |
6 |
1 |
61 |
461.63 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.76 |
9.64 |
-12.67 |
1 |
6 |
0 |
59 |
460.622 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.76 |
11.36 |
-37.79 |
2 |
6 |
1 |
61 |
461.63 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.34 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
11.76 |
-30.91 |
2 |
6 |
1 |
61 |
461.63 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.76 |
10.76 |
-13.51 |
1 |
6 |
0 |
59 |
460.622 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.76 |
11.6 |
-41.31 |
2 |
6 |
1 |
61 |
461.63 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.30 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.09 |
12.57 |
-38.13 |
2 |
5 |
1 |
51 |
481.611 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.09 |
11.77 |
-10.31 |
1 |
5 |
0 |
50 |
480.603 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.09 |
11.91 |
-33.93 |
2 |
5 |
1 |
51 |
481.611 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.32 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
10.49 |
-27.52 |
1 |
7 |
0 |
80 |
479.631 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OX1R-1-E |
Orexin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.34 |
Functional ≤ 10μM
|
|
OX2R-1-E |
Orexin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
12.02 |
-21.06 |
1 |
7 |
0 |
80 |
491.642 |
5 |
↓
|
|