|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 86 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AK1C3-1-E |
Aldo-keto-reductase Family 1 Member C3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2600 |
0.41 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.58 |
Binding ≤ 10μM
|
|
FABPL-2-E |
Fatty Acid-binding Protein, Liver (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3220 |
0.40 |
Binding ≤ 10μM
|
|
IL8-1-E |
Interleukin-8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.60 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
70 |
0.53 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
160 |
0.50 |
Binding ≤ 10μM
|
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
77 |
0.52 |
Binding ≤ 10μM
|
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
2500 |
0.41 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
8.81 |
-48.12 |
1 |
3 |
-1 |
52 |
295.145 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5550 |
0.35 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8950 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
-0.13 |
-53.75 |
2 |
6 |
-1 |
107 |
307.303 |
4 |
↓
|
|
|
|
Analogs
-
22066291
-
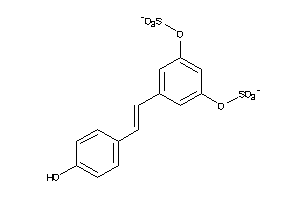
-
22066296
-
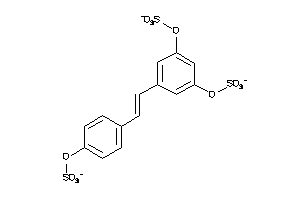
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CISD1-1-E |
CDGSH Iron-sulfur Domain-containing Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3360 |
0.36 |
Binding ≤ 10μM |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3600 |
0.36 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7530 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
-0.09 |
-48.08 |
2 |
6 |
-1 |
107 |
307.303 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
140 |
0.37 |
Binding ≤ 10μM
|
|
CYNT-1-B |
Carbonic Anhydrase (cluster #1 Of 3), Bacterial |
Bacteria |
713 |
0.33 |
Binding ≤ 10μM
|
|
B5SU02-2-E |
Alpha Carbonic Anhydrase (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
34 |
0.40 |
Binding ≤ 10μM
|
|
C0IX24-1-E |
Carbonic Anhydrase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
690 |
0.33 |
Binding ≤ 10μM
|
|
CAH12-1-E |
Carbonic Anhydrase XII (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
18 |
0.42 |
Binding ≤ 10μM |
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.38 |
Binding ≤ 10μM
|
|
CAH14-1-E |
Carbonic Anhydrase XIV (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
689 |
0.33 |
Binding ≤ 10μM
|
|
CAH15-2-E |
Carbonic Anhydrase 15 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
45 |
0.40 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Binding ≤ 10μM |
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 16), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-1-E |
Carbonic Anhydrase VA (cluster #1 Of 10), Eukaryotic |
Eukaryotes |
794 |
0.33 |
Binding ≤ 10μM
|
|
CAH5B-1-E |
Carbonic Anhydrase VB (cluster #1 Of 9), Eukaryotic |
Eukaryotes |
93 |
0.38 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
94 |
0.38 |
Binding ≤ 10μM
|
|
CAH7-1-E |
Carbonic Anhydrase VII (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2170 |
0.30 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM |
|
COX2-1-E |
Cytochrome C Oxidase Subunit 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
60 |
0.39 |
Binding ≤ 10μM
|
|
MK14-1-E |
MAP Kinase P38 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
810 |
0.33 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9730 |
0.27 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9 |
0.43 |
Binding ≤ 10μM
|
|
Q8HZR1-1-E |
Cyclooxygenase-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5570 |
0.28 |
Binding ≤ 10μM
|
|
Q8SPQ9-2-E |
Cyclooxygenase-2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.33 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Functional ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.29 |
Functional ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.27 |
ADME/T ≤ 10μM
|
|
CAN-1-F |
Carbonic Anhydrase (cluster #1 Of 3), Fungal |
Fungi |
108 |
0.38 |
Binding ≤ 10μM
|
|
Q5AJ71-1-F |
Carbonic Anhydrase (cluster #1 Of 4), Fungal |
Fungi |
21 |
0.41 |
Binding ≤ 10μM
|
|
Z100741-1-O |
MC9 (Mast Cells) (cluster #1 Of 2), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
6670 |
0.28 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
5000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
5.33 |
-11.94 |
2 |
5 |
0 |
78 |
381.379 |
4 |
↓
|
|
|
|
Analogs
-
3978494
-
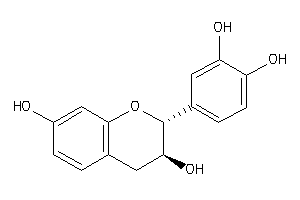
-
8681596
-
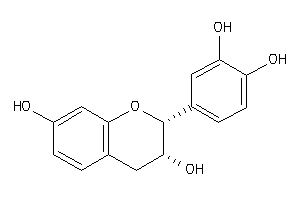
-
8681597
-
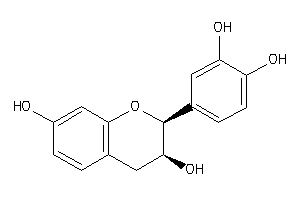
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4720 |
0.36 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5030 |
0.35 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2500 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
-4.3 |
-13.35 |
5 |
6 |
0 |
110 |
290.271 |
1 |
↓
|
|
|
|
Analogs
-
34395899
-
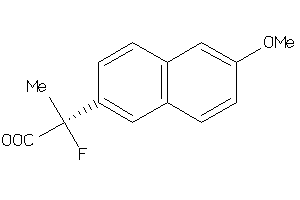
Draw
Identity
99%
90%
80%
70%
Vendors
And 89 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
7.87 |
-47.82 |
0 |
3 |
-1 |
49 |
229.255 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
990 |
0.37 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
83 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
680 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
4.02 |
-13.34 |
2 |
4 |
0 |
62 |
266.659 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.48 |
1.98 |
-41.28 |
1 |
4 |
-1 |
65 |
265.651 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2700 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
4.47 |
-12.72 |
2 |
4 |
0 |
62 |
283.114 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.00 |
2.43 |
-40.92 |
1 |
4 |
-1 |
65 |
282.106 |
2 |
↓
|
|
|
|
Analogs
-
40117605
-
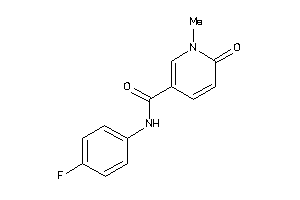
-
40117644
-
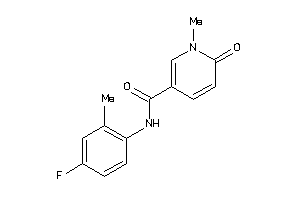
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3500 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
3.51 |
-13.33 |
2 |
4 |
0 |
62 |
232.214 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-3-E |
Cyclooxygenase-1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
9200 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
3.91 |
-15.17 |
2 |
4 |
0 |
62 |
248.669 |
2 |
↓
|
|
|
|
Analogs
-
12353732
-
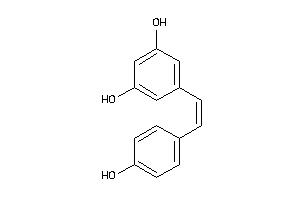
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
169 |
0.56 |
Binding ≤ 10μM
|
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2210 |
0.47 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
950 |
0.50 |
Binding ≤ 10μM
|
|
CAH13-7-E |
Carbonic Anhydrase XIII (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
4090 |
0.44 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9360 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
2770 |
0.46 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9090 |
0.42 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4470 |
0.44 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
4750 |
0.44 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4640 |
0.44 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8070 |
0.42 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4350 |
0.44 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
810 |
0.50 |
Binding ≤ 10μM
|
|
ESR1-5-E |
Estrogen Receptor Alpha (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
785 |
0.50 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.50 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3490 |
0.45 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2840 |
0.46 |
Functional ≤ 10μM
|
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
6918 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
5000 |
0.44 |
Functional ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 9), Other |
Other |
4000 |
0.44 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
0.79 |
-8.46 |
3 |
3 |
0 |
61 |
228.247 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
6.6 |
-5.41 |
2 |
2 |
0 |
40 |
266.34 |
5 |
↓
|
|
|
|
Analogs
-
5760137
-
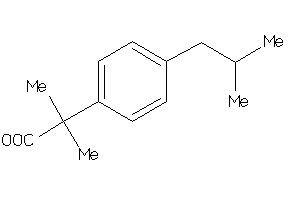
-
5760156
-
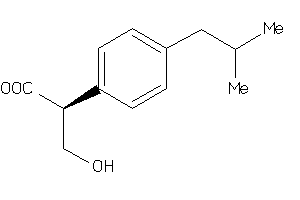
-
5760166
-
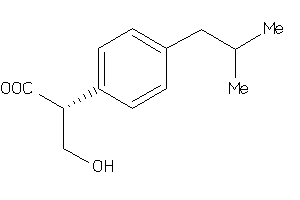
-
2647
-
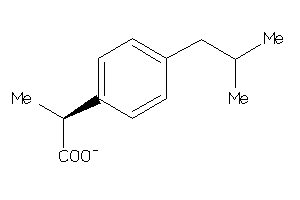
Draw
Identity
99%
90%
80%
70%
Vendors
And 94 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALBU-1-E |
Serum Albumin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2600 |
0.52 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM |
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1500 |
0.54 |
Binding ≤ 10μM
|
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.50 |
Binding ≤ 10μM
|
|
S22AK-1-E |
Solute Carrier Family 22 Member 20 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1122 |
0.56 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.65 |
Functional ≤ 10μM
|
|
CXCR2-1-E |
C-X-C Chemokine Receptor Type 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.65 |
Functional ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
9.06 |
-46.94 |
0 |
2 |
-1 |
40 |
205.277 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR44-1-E |
G Protein-coupled Receptor 44 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.32 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
50 |
0.43 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
100 |
0.41 |
Binding ≤ 10μM
|
|
PPARG-2-E |
Peroxisome Proliferator-activated Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
13.18 |
-53.63 |
0 |
2 |
-1 |
40 |
339.411 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4720 |
0.36 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5030 |
0.35 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5300 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
-4.98 |
-12.32 |
5 |
6 |
0 |
110 |
290.271 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9200 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
4.28 |
-9.11 |
2 |
4 |
0 |
63 |
296.779 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3200 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
4.28 |
-10.04 |
2 |
4 |
0 |
63 |
296.779 |
3 |
↓
|
|
|
|
Analogs
-
391883
-
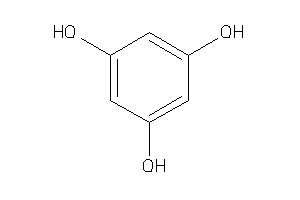
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
7500 |
0.90 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
7700 |
0.89 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
8700 |
0.89 |
Binding ≤ 10μM
|
|
CAH5B-8-E |
Carbonic Anhydrase VB (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
7100 |
0.90 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3600 |
0.95 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6420 |
0.91 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
-1.22 |
-5.57 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.32 |
1.74 |
-46.85 |
0 |
2 |
-1 |
40 |
109.104 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
6.59 |
-56.84 |
0 |
4 |
-1 |
66 |
179.151 |
3 |
↓
|
|
|
|
Analogs
-
2167188
-
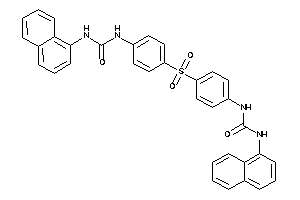
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9200 |
0.35 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
170 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
3.2 |
-22.27 |
2 |
5 |
0 |
75 |
290.344 |
3 |
↓
|
|
|
|
Analogs
-
34592840
-
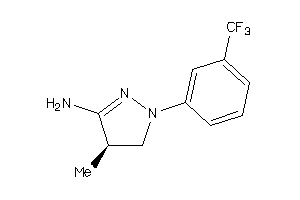
-
34592841
-
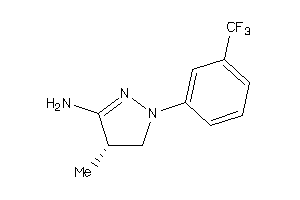
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CLTR2-1-E |
Cysteinyl Leukotriene Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
700 |
0.54 |
Binding ≤ 10μM
|
|
LOX5-5-E |
Arachidonate 5-lipoxygenase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
890 |
0.53 |
Binding ≤ 10μM
|
|
LT4R1-1-E |
Leukotriene B4 Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.54 |
Binding ≤ 10μM
|
|
PGH1-3-E |
Cyclooxygenase-1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
800 |
0.53 |
Binding ≤ 10μM |
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
1300 |
0.52 |
Binding ≤ 10μM |
|
LOX5-3-E |
Arachidonate 5-lipoxygenase (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
750 |
0.54 |
Functional ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
8500 |
0.44 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
1.29 |
-9.17 |
2 |
3 |
0 |
42 |
229.205 |
2 |
↓
|
|
|
|
Analogs
-
6322373
-
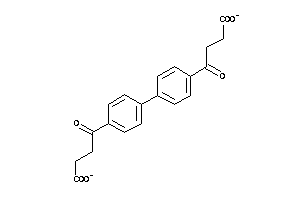
-
26544446
-
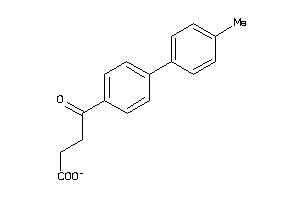
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3900 |
0.40 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8100 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
9.87 |
-46.3 |
0 |
3 |
-1 |
57 |
253.277 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
590 |
0.30 |
Binding ≤ 10μM
|
|
PGH2-8-E |
Cyclooxygenase-2 (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
270 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
7.23 |
-13.12 |
2 |
6 |
0 |
81 |
414.889 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
26544397
-
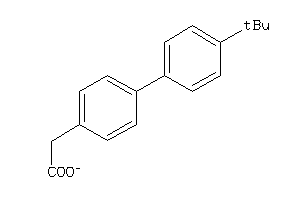
-
1257461
-
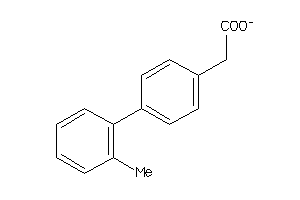
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
32 |
0.62 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
21 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
8.84 |
-48.82 |
0 |
2 |
-1 |
40 |
225.267 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.47 |
-1.1 |
-13.63 |
2 |
4 |
0 |
74 |
318.328 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
870 |
0.37 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
7000 |
0.31 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
9.09 |
-11.15 |
0 |
2 |
0 |
34 |
411.317 |
3 |
↓
|
|
|
|
Analogs
-
5760137
-

-
5760156
-

-
5760166
-

-
2647
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 77 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALBU-1-E |
Serum Albumin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
IL8-1-E |
Interleukin-8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.68 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1100 |
0.56 |
Binding ≤ 10μM |
|
S22A6-1-E |
Solute Carrier Family 22 Member 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4700 |
0.50 |
Binding ≤ 10μM
|
|
S22AK-1-E |
Solute Carrier Family 22 Member 20 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1122 |
0.56 |
Binding ≤ 10μM
|
|
CXCR1-1-E |
Interleukin-8 Receptor A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Functional ≤ 10μM
|
|
CXCR2-1-E |
C-X-C Chemokine Receptor Type 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Functional ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
9.02 |
-45.75 |
0 |
2 |
-1 |
40 |
205.277 |
4 |
↓
|
|
|
|
Analogs
-
8667
-
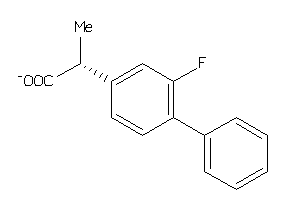
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
10 |
-44.65 |
0 |
2 |
-1 |
40 |
243.257 |
3 |
↓
|
|
|
|
Analogs
-
323
-
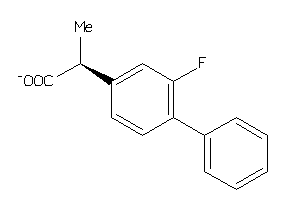
Draw
Identity
99%
90%
80%
70%
Vendors
And 53 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABPL-3-E |
Fatty Acid-binding Protein, Liver (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1180 |
0.46 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
170 |
0.53 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
500 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.05 |
9.98 |
-43.92 |
0 |
2 |
-1 |
40 |
243.257 |
3 |
↓
|
|
|
|
Analogs
-
34007650
-
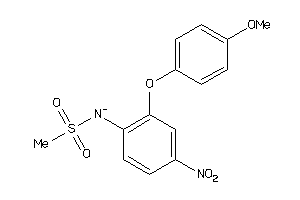
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3760 |
0.36 |
Binding ≤ 10μM
|
|
PGH2-8-E |
Cyclooxygenase-2 (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
400 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
4.93 |
-37.19 |
0 |
7 |
-1 |
103 |
307.307 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.81 |
4.82 |
-11.73 |
1 |
7 |
0 |
101 |
308.315 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 43 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4720 |
0.36 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5030 |
0.35 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
-4.31 |
-13.19 |
5 |
6 |
0 |
110 |
290.271 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.29 |
Binding ≤ 10μM
|
|
GPR44-1-E |
G Protein-coupled Receptor 44 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.29 |
Binding ≤ 10μM
|
|
IL8-1-E |
Interleukin-8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.41 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM |
|
PGH2-8-E |
Cyclooxygenase-2 (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
9 |
0.45 |
Binding ≤ 10μM
|
|
PTGDS-1-E |
Prostaglandin-H2 D-isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.35 |
Binding ≤ 10μM
|
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7000 |
0.29 |
Functional ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Functional ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.43 |
Functional ≤ 10μM
|
|
THAS-1-E |
Thromboxane-A Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.39 |
Functional ≤ 10μM
|
|
Z102213-1-O |
Blood (cluster #1 Of 2), Other |
Other |
400 |
0.36 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
800 |
0.34 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
500 |
0.35 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
800 |
0.34 |
Functional ≤ 10μM
|
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
53 |
0.41 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
4 |
0.47 |
Functional ≤ 10μM
|
|
Z81267-1-O |
Mononuclear Cell Line (cluster #1 Of 1), Other |
Other |
300 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
11.18 |
-50.91 |
0 |
5 |
-1 |
71 |
356.785 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
1.99 |
-14.13 |
3 |
6 |
0 |
98 |
397.378 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
58 |
0.53 |
Binding ≤ 10μM
|
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
35 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
8.86 |
-49.89 |
1 |
3 |
-1 |
52 |
274.727 |
4 |
↓
|
|