|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
11.08 |
-11.14 |
1 |
5 |
0 |
65 |
361.463 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.37 |
11.08 |
-11.88 |
1 |
5 |
0 |
65 |
361.463 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
0.71 |
-10.43 |
0 |
3 |
0 |
33 |
352.865 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.33 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.29 |
Binding ≤ 10μM |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
14 |
0.31 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
56 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
-3.66 |
-17.93 |
1 |
9 |
0 |
109 |
510.605 |
12 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
14243647
-
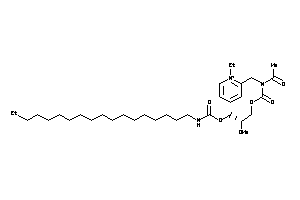
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.27 |
Binding ≤ 10μM |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.23 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
55 |
0.24 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
12 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
18.67 |
-37.1 |
1 |
9 |
1 |
98 |
606.869 |
27 |
↓
|
|
|
|
Analogs
-
14243646
-
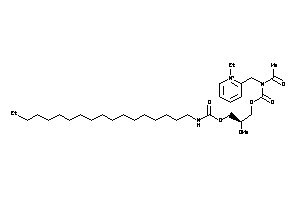
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.27 |
Binding ≤ 10μM |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.23 |
Functional ≤ 10μM
|
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
91 |
0.23 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
55 |
0.24 |
Functional ≤ 10μM |
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
12 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
18.64 |
-35.54 |
1 |
9 |
1 |
98 |
606.869 |
27 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
9.62 |
-18 |
0 |
7 |
0 |
73 |
455.971 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
10.55 |
-40.91 |
1 |
7 |
1 |
74 |
456.979 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.16 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.26 |
Binding ≤ 10μM |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.26 |
Functional ≤ 10μM
|
|
Z50588-2-O |
Canis Familiaris (cluster #2 Of 7), Other |
Other |
6600 |
0.16 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
29 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
16.7 |
-28.15 |
2 |
9 |
0 |
111 |
605.098 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6800 |
0.16 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.26 |
Binding ≤ 10μM |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.26 |
Functional ≤ 10μM
|
|
Z50588-2-O |
Canis Familiaris (cluster #2 Of 7), Other |
Other |
6600 |
0.16 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
29 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
16.55 |
-28.49 |
2 |
9 |
0 |
111 |
605.098 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
1532042
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
8.54 |
-11 |
0 |
5 |
0 |
46 |
372.461 |
6 |
↓
|
|
|
|
Analogs
-
8195613
-
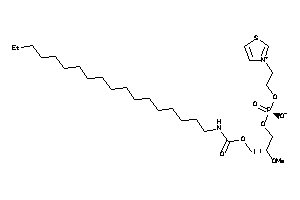
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.22 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
16.47 |
-52.88 |
1 |
9 |
0 |
110 |
592.78 |
28 |
↓
|
|
|
|
Analogs
-
8034876
-
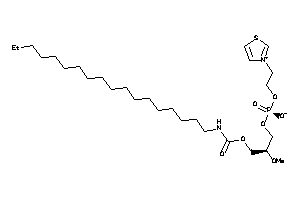
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.22 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.52 |
16.48 |
-53.33 |
1 |
9 |
0 |
110 |
592.78 |
28 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.22 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.22 |
Binding ≤ 10μM
|
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7800 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3250 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
12.23 |
-10.38 |
1 |
3 |
0 |
33 |
433.567 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3250 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
11.95 |
-9.65 |
1 |
3 |
0 |
33 |
433.567 |
6 |
↓
|
|
|
|
|
|
|
Analogs
-
6145655
-
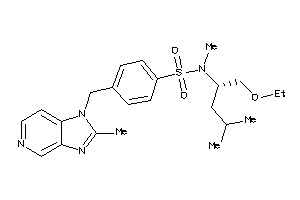
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 8), Other |
Other |
4 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
10.55 |
-17.52 |
0 |
8 |
0 |
94 |
458.584 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.28 |
11.07 |
-43.58 |
1 |
8 |
1 |
96 |
459.592 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
180 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
7.12 |
-14.58 |
1 |
5 |
0 |
63 |
368.849 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
8.92 |
-10.07 |
0 |
4 |
0 |
43 |
393.697 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.17 |
9.86 |
-33.76 |
1 |
4 |
1 |
45 |
394.705 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.46 |
14.21 |
-22.36 |
0 |
7 |
0 |
73 |
479.515 |
4 |
↓
|
|
|
|
Analogs
-
1532042
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.28 |
Binding ≤ 10μM
|
|
Z50466-1-O |
Trypanosoma Cruzi (cluster #1 Of 8), Other |
Other |
4400 |
0.28 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
7700 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
8.79 |
-11.19 |
0 |
5 |
0 |
46 |
372.461 |
6 |
↓
|
|
|
|
|
|
|
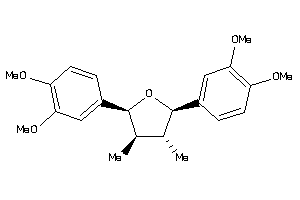
Analogs
-
1532042
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(2S,3R,4S,5R)-2,5-bis(3,4-dimethoxyphenyl)-3,4-dimethyl-tetrahydrofuran
(2S,3R,4S,5R)-2,5-bis(3,4-dimeth…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
8.27 |
-11.02 |
0 |
5 |
0 |
46 |
372.461 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.72 |
1.24 |
-43.61 |
1 |
3 |
1 |
30 |
416.976 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.32 |
Binding ≤ 10μM |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.32 |
Functional ≤ 10μM |
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 1), Other |
Other |
4 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.28 |
2.16 |
-16.31 |
0 |
6 |
0 |
60 |
510.594 |
4 |
↓
|
|
|
|
Analogs
-
8860523
-
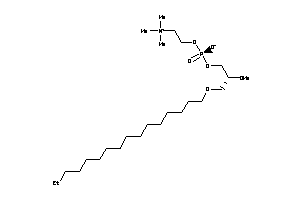
-
14255579
-
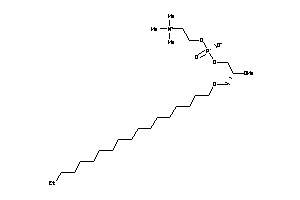
-
17654196
-
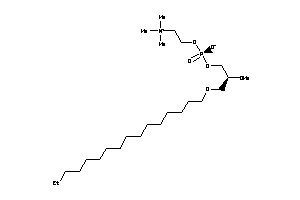
-
38143169
-
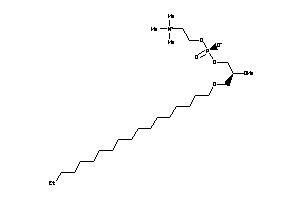
-
43379726
-
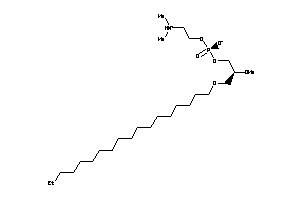
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.23 |
Functional ≤ 10μM
|
|
Z50417-1-O |
Leishmania Infantum (cluster #1 Of 4), Other |
Other |
820 |
0.24 |
Functional ≤ 10μM |
|
Z50607-9-O |
Human Immunodeficiency Virus 1 (cluster #9 Of 10), Other |
Other |
920 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
260 |
0.26 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
6100 |
0.21 |
Functional ≤ 10μM
|
|
Z80166-2-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #2 Of 12), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM |
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 11), Other |
Other |
6100 |
0.21 |
Functional ≤ 10μM
|
|
Z80678-1-O |
AMML Cell Line (cluster #1 Of 1), Other |
Other |
1350 |
0.23 |
Functional ≤ 10μM
|
|
Z80678-1-O |
AMML Cell Line (cluster #1 Of 1), Other |
Other |
3300 |
0.22 |
Functional ≤ 10μM
|
|
Z80777-1-O |
CML/BC Cell Line (cluster #1 Of 1), Other |
Other |
250 |
0.26 |
Functional ≤ 10μM
|
|
Z80777-1-O |
CML/BC Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.23 |
Functional ≤ 10μM
|
|
Z81245-2-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
3000 |
0.22 |
Functional ≤ 10μM
|
|
Z81252-2-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #2 Of 11), Other |
Other |
5000 |
0.21 |
Functional ≤ 10μM |
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
4960 |
0.21 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
18.9 |
-48.04 |
0 |
7 |
0 |
77 |
523.736 |
27 |
↓
|
|
|
|
Analogs
-
17654196
-

-
38143169
-

-
43379726
-

-
43379728
-
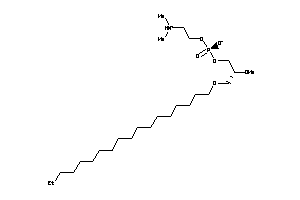
-
8195621
-
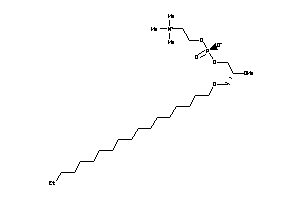
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-2-E |
Platelet Activating Factor Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.23 |
Functional ≤ 10μM
|
|
Z50417-1-O |
Leishmania Infantum (cluster #1 Of 4), Other |
Other |
820 |
0.24 |
Functional ≤ 10μM |
|
Z50607-9-O |
Human Immunodeficiency Virus 1 (cluster #9 Of 10), Other |
Other |
920 |
0.24 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
260 |
0.26 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
6100 |
0.21 |
Functional ≤ 10μM
|
|
Z80166-2-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #2 Of 12), Other |
Other |
2000 |
0.23 |
Functional ≤ 10μM |
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 11), Other |
Other |
6100 |
0.21 |
Functional ≤ 10μM
|
|
Z80678-1-O |
AMML Cell Line (cluster #1 Of 1), Other |
Other |
1350 |
0.23 |
Functional ≤ 10μM
|
|
Z80678-1-O |
AMML Cell Line (cluster #1 Of 1), Other |
Other |
3300 |
0.22 |
Functional ≤ 10μM
|
|
Z80777-1-O |
CML/BC Cell Line (cluster #1 Of 1), Other |
Other |
250 |
0.26 |
Functional ≤ 10μM
|
|
Z80777-1-O |
CML/BC Cell Line (cluster #1 Of 1), Other |
Other |
1500 |
0.23 |
Functional ≤ 10μM
|
|
Z81245-2-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
3000 |
0.22 |
Functional ≤ 10μM
|
|
Z81252-2-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #2 Of 11), Other |
Other |
5000 |
0.21 |
Functional ≤ 10μM |
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
4960 |
0.21 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
520 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
10.82 |
-10.95 |
0 |
4 |
0 |
36 |
385.511 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.18 |
13.32 |
-116.02 |
2 |
4 |
2 |
39 |
387.527 |
6 |
↓
|
|
|
|
Analogs
-
13729124
-
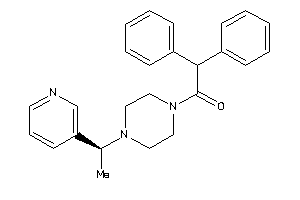
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.65 |
-11.29 |
0 |
4 |
0 |
36 |
385.511 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.47 |
12.92 |
-110.83 |
2 |
4 |
2 |
39 |
387.527 |
5 |
↓
|
|
|
|
Analogs
-
13729121
-
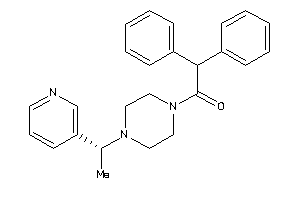
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
10.57 |
-9.57 |
0 |
4 |
0 |
36 |
385.511 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.47 |
12.8 |
-113.59 |
2 |
4 |
2 |
39 |
387.527 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
9.76 |
-12.17 |
0 |
4 |
0 |
36 |
371.484 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5200 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.25 |
-0.25 |
-14.67 |
0 |
8 |
0 |
81 |
385.42 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
11.16 |
-16.4 |
0 |
5 |
0 |
54 |
399.494 |
5 |
↓
|
|
|
|
Analogs
-
12951351
-
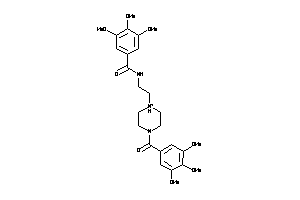
-
19419633
-
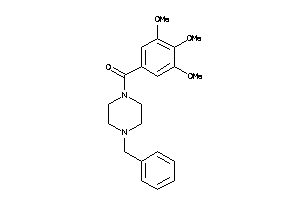
-
19555329
-
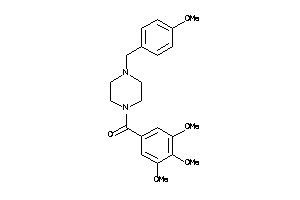
-
19565297
-
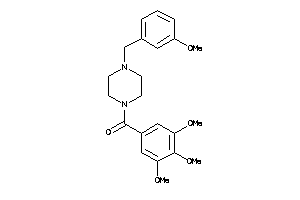
-
19608260
-
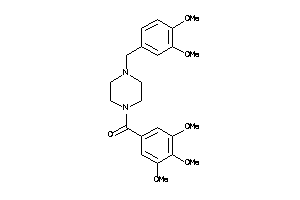
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-3-E |
Platelet Activating Factor Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9400 |
0.21 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1780 |
0.24 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
1.47 |
-18.28 |
0 |
10 |
0 |
96 |
474.51 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.28 |
16.03 |
-15.69 |
0 |
6 |
0 |
60 |
510.594 |
4 |
↓
|
|