|
|
Analogs
-
2172695
-
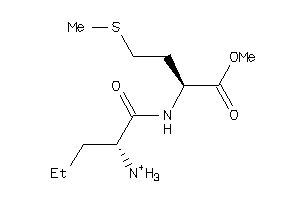
-
2172698
-
-
2172700
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.45 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.35 |
-3.97 |
-76.43 |
4 |
5 |
0 |
96 |
280.415 |
9 |
↓
|
|
|
|
Analogs
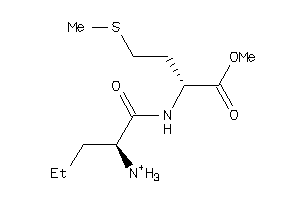
-
33943584
-
-
35024386
-
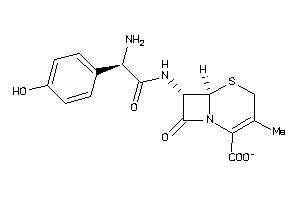
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.31 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.96 |
1.55 |
-61.03 |
4 |
8 |
-1 |
136 |
362.387 |
4 |
↓
|
|
|
|
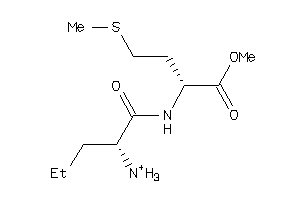
Analogs
-
32211233
-
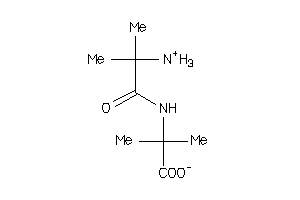
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.66 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.26 |
-3.82 |
-63.85 |
4 |
5 |
0 |
96 |
160.173 |
3 |
↓
|
|
|
|
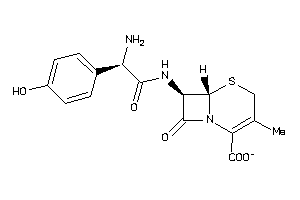
Analogs
-
2384984
-
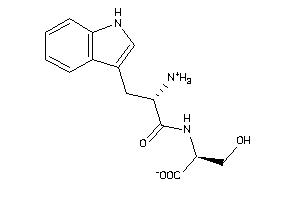
-
44624305
-
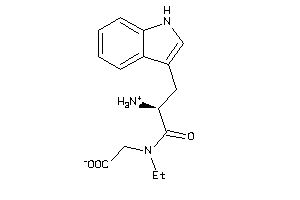
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Oligopeptide Transporter, Kidney Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
-4.39 |
-58.72 |
5 |
6 |
0 |
112 |
275.308 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.61 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.53 |
-6.66 |
-72.55 |
5 |
6 |
0 |
117 |
176.172 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.60 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.53 |
-6.73 |
-59.69 |
5 |
6 |
0 |
117 |
176.172 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
2583985
-
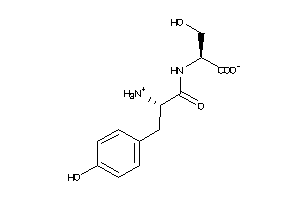
-
4787311
-
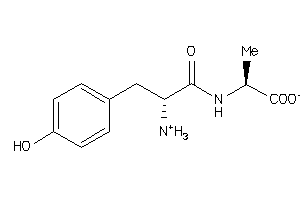
-
4787313
-
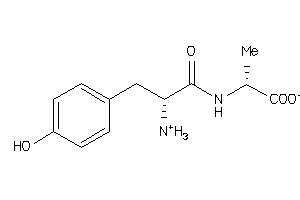
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.42 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.27 |
-5.43 |
-63.57 |
5 |
6 |
0 |
117 |
252.27 |
5 |
↓
|
|
|
|
Analogs
-
15721974
-
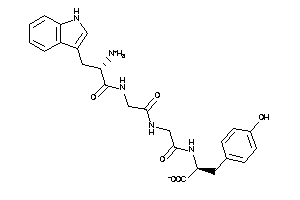
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.26 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.05 |
2.11 |
-72.15 |
7 |
9 |
0 |
162 |
424.457 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.21 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.01 |
-6.74 |
-68.15 |
8 |
10 |
0 |
173 |
576.657 |
11 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAN1-1-E |
Calpain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.59 |
Binding ≤ 10μM
|
|
CPNS1-1-E |
Calpain Small Subunit 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.59 |
Binding ≤ 10μM
|
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.40 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.01 |
-2.02 |
-70.32 |
4 |
5 |
0 |
96 |
264.325 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S15A2-1-E |
Solute Carrier Family 15 Member 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.72 |
ADME/T ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.04 |
-4.49 |
-63.15 |
4 |
5 |
0 |
96 |
146.146 |
3 |
↓
|
|