|
|
Analogs
-
20995032
-
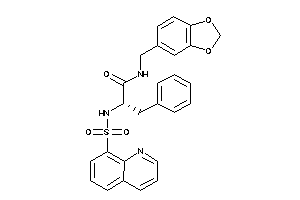
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
2 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
7.37 |
-23.51 |
2 |
8 |
0 |
107 |
489.553 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.15 |
6.79 |
-67.02 |
1 |
8 |
-1 |
109 |
488.545 |
8 |
↓
|
|
|
|
Analogs
-
20995028
-
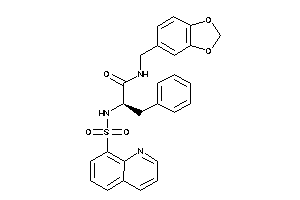
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
2 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
7.46 |
-22.85 |
2 |
8 |
0 |
107 |
489.553 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.15 |
7 |
-62.49 |
1 |
8 |
-1 |
109 |
488.545 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
1 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.45 |
12.67 |
-52.86 |
2 |
6 |
1 |
78 |
482.01 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.91 |
9.81 |
-15.9 |
1 |
6 |
0 |
76 |
481.002 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.45 |
11.65 |
-27.88 |
1 |
6 |
0 |
76 |
481.002 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
5793773
-
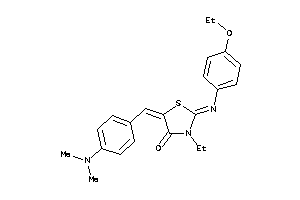
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
24 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
10.69 |
-9.27 |
0 |
5 |
0 |
47 |
395.528 |
6 |
↓
|
|
|
|
Analogs
-
8816253
-
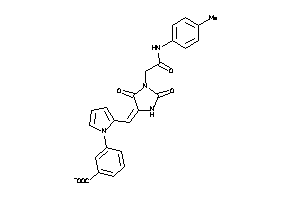
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
18 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
11 |
-63.88 |
2 |
9 |
-1 |
129 |
443.439 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
605 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
605 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
30 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
8.95 |
-53.08 |
0 |
7 |
-1 |
93 |
431.877 |
5 |
↓
|
|
|
|
Analogs
-
88758
-
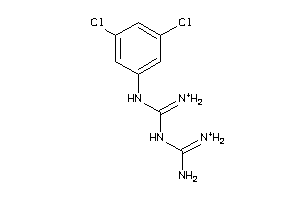
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
6.42 |
-40 |
7 |
5 |
1 |
100 |
247.109 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.61 |
6.55 |
-5.63 |
6 |
5 |
0 |
98 |
246.101 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.61 |
6.44 |
-5.23 |
6 |
5 |
0 |
98 |
246.101 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA5-2-E |
Fibronectin Receptor Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.26 |
Binding ≤ 10μM
|
|
ITAV-1-E |
Vitronectin Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.29 |
Binding ≤ 10μM
|
|
ITB5-1-E |
Integrin Beta-5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.29 |
Binding ≤ 10μM
|
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
19 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.75 |
-11.08 |
-77.02 |
9 |
15 |
0 |
240 |
588.666 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.16 |
Binding ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.16 |
Binding ≤ 10μM
|
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
210 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.75 |
-2.85 |
-194.48 |
17 |
22 |
1 |
385 |
716.774 |
24 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.77 |
-3.21 |
-154.73 |
16 |
22 |
0 |
386 |
715.766 |
23 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.75 |
-3.25 |
-155.07 |
16 |
22 |
0 |
384 |
715.766 |
24 |
↓
|
|