|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
240 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.44 |
10.43 |
-8.29 |
0 |
2 |
0 |
20 |
279.383 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50142-1-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #1 Of 2), Other |
Other |
1860 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.08 |
9.94 |
-9.62 |
0 |
2 |
0 |
20 |
285.774 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
2100 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
1.49 |
-6.97 |
0 |
2 |
0 |
20 |
257.377 |
2 |
↓
|
|
|
|
Analogs
-
11592802
-
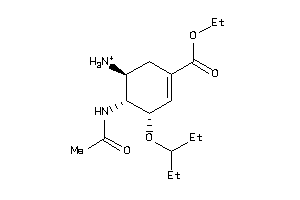
-
36451498
-
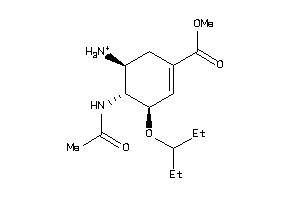
-
3874571
-
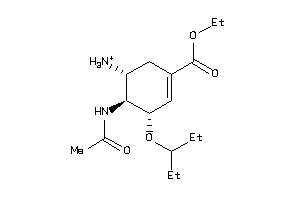
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101821-1-O |
H2N2 Subtype (cluster #1 Of 3), Other |
Other |
1450 |
0.37 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
260 |
0.42 |
Functional ≤ 10μM
|
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
8600 |
0.32 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
2600 |
0.36 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
A5Z252-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
8 |
0.52 |
Binding ≤ 10μM
|
|
C4LRQ6-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.57 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
2 |
0.55 |
Binding ≤ 10μM
|
|
Q2LFS1-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
5 |
0.53 |
Binding ≤ 10μM
|
|
Q6Q793-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
5.84 |
-62.6 |
4 |
6 |
1 |
92 |
313.418 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.85 |
5.55 |
-12.89 |
3 |
6 |
0 |
91 |
312.41 |
8 |
↓
|
|
|
|
Analogs
-
4245717
-
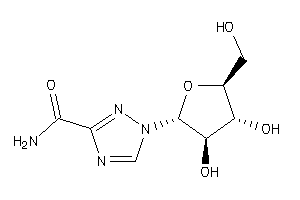
-
6603353
-
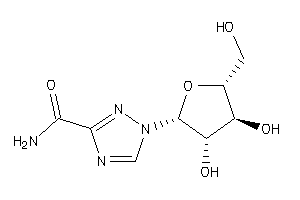
Draw
Identity
99%
90%
80%
70%
Vendors
-
-
-
- API0004062
- API0009704
- CCH0012019
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100497-1-O |
Bovine Viral Diarrhea Virus (cluster #1 Of 3), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z100811-1-O |
Influenza A Virus (H5N1) (cluster #1 Of 1), Other |
Other |
9400 |
0.41 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
9000 |
0.42 |
Functional ≤ 10μM
|
|
Z50142-1-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #1 Of 2), Other |
Other |
9700 |
0.41 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
9000 |
0.42 |
Functional ≤ 10μM
|
|
Z50525-1-O |
Tacaribe Virus (cluster #1 Of 1), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
8300 |
0.42 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z50641-1-O |
West Nile Virus (cluster #1 Of 2), Other |
Other |
1100 |
0.49 |
Functional ≤ 10μM
|
|
Z50651-1-O |
Vesicular Stomatitis Virus (cluster #1 Of 2), Other |
Other |
10000 |
0.41 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
9800 |
0.41 |
Functional ≤ 10μM |
|
Z50678-2-O |
Respiratory Syncytial Virus (cluster #2 Of 2), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
1100 |
0.49 |
Functional ≤ 10μM
|
|
Z80249-1-O |
MDCK (Kidney Cells) (cluster #1 Of 2), Other |
Other |
4900 |
0.44 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
5700 |
0.43 |
Functional ≤ 10μM |
|
Z80823-1-O |
E6SM (Embryonic Skin-muscle Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
5 |
0.68 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1500 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.77 |
-8.13 |
-17.41 |
5 |
9 |
0 |
144 |
244.207 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM |
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM |
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM |
|
NEUR3-1-E |
Sialidase-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.31 |
Binding ≤ 10μM
|
|
NEUR4-1-E |
Sialidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.34 |
Binding ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Binding ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 1), Other |
Other |
5 |
0.51 |
Binding ≤ 10μM
|
|
Z100811-1-O |
Influenza A Virus (H5N1) (cluster #1 Of 1), Other |
Other |
2800 |
0.34 |
Functional ≤ 10μM
|
|
Z101821-1-O |
H2N2 Subtype (cluster #1 Of 3), Other |
Other |
760 |
0.37 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
680 |
0.38 |
Functional ≤ 10μM
|
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
11 |
0.48 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
90 |
0.43 |
Functional ≤ 10μM
|
|
Z80249-1-O |
MDCK (Kidney Cells) (cluster #1 Of 2), Other |
Other |
40 |
0.45 |
Functional ≤ 10μM |
|
A5Z252-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
|
B4URF0-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
4 |
0.51 |
Binding ≤ 10μM
|
|
C4LRQ6-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
3 |
0.52 |
Binding ≤ 10μM
|
|
Q2LFS1-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
|
Q6Q793-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
1 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.64 |
-6.67 |
-67.62 |
9 |
11 |
0 |
205 |
332.313 |
6 |
↓
|
|
|
|
Analogs
-
3978503
-
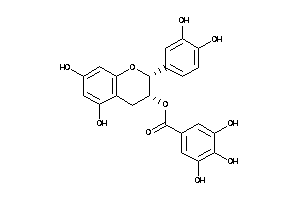
-
4534390
-
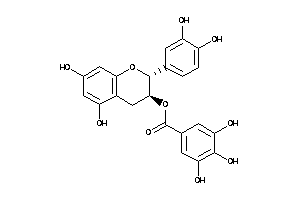
-
4544252
-
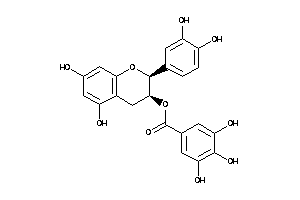
-
8681494
-
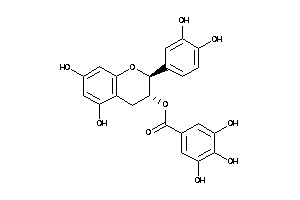
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
6PGD-1-E |
6-phosphogluconate Dehydrogenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
720 |
0.26 |
Binding ≤ 10μM
|
|
BCL2-1-E |
Apoptosis Regulator Bcl-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
234 |
0.28 |
Binding ≤ 10μM
|
|
ERG1-3-E |
Squalene Monooxygenase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
690 |
0.26 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6026 |
0.22 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.23 |
Binding ≤ 10μM
|
|
MK14-2-E |
MAP Kinase P38 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2210 |
0.24 |
Binding ≤ 10μM
|
|
MMP14-1-E |
Matrix Metalloproteinase 14 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6800 |
0.22 |
Binding ≤ 10μM
|
|
MMP2-2-E |
72 KDa Type IV Collagenase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9600 |
0.21 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.34 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.28 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.27 |
Binding ≤ 10μM
|
|
G6PD-1-F |
Glucose-6-phosphate 1-dehydrogenase (cluster #1 Of 1), Fungal |
Fungi |
250 |
0.28 |
Binding ≤ 10μM
|
|
Z50142-1-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #1 Of 2), Other |
Other |
391 |
0.27 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
9900 |
0.21 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
9400 |
0.21 |
Functional ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
730 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
-6.71 |
-20.66 |
8 |
11 |
0 |
197 |
458.375 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.25 |
-5.81 |
-60.24 |
7 |
11 |
-1 |
200 |
457.367 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.25 |
-5.67 |
-56.02 |
7 |
11 |
-1 |
200 |
457.367 |
4 |
↓
|
|
|
|
Analogs
-
3831612
-
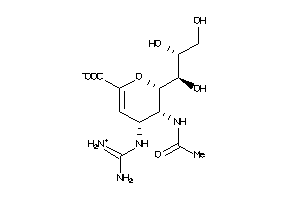
-
3831613
-
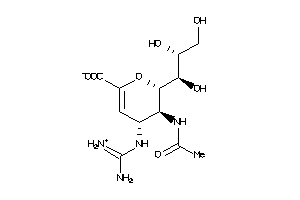
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.11 |
-10.28 |
-72.57 |
8 |
11 |
0 |
191 |
346.34 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.43 |
-10.24 |
-53.08 |
7 |
12 |
-1 |
209 |
365.315 |
7 |
↓
|
|
|
|
Analogs
-
6777826
-
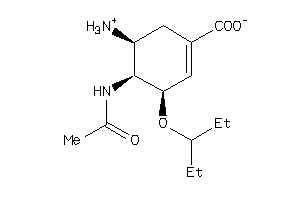
-
6777828
-
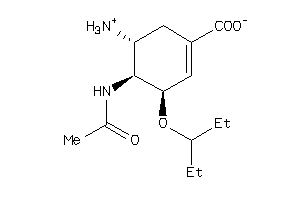
-
6777829
-
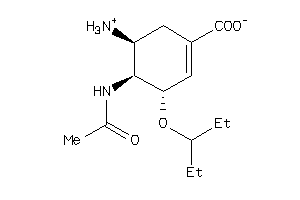
-
6777830
-
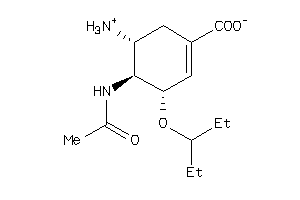
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
A8K327-1-E |
Sialidase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
NEUR1-1-E |
Sialidase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.63 |
Binding ≤ 10μM
|
|
NEUR2-1-E |
Sialidase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
NEUR4-1-E |
Sialidase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Binding ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
4 |
0.59 |
Binding ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 1), Other |
Other |
2 |
0.61 |
Binding ≤ 10μM
|
|
Z101821-1-O |
H2N2 Subtype (cluster #1 Of 3), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z101822-1-O |
H3N2 Subtype (cluster #1 Of 2), Other |
Other |
1 |
0.63 |
Functional ≤ 10μM
|
|
Z50142-2-O |
Influenza A Virus (A/PR/8/34(H1N1)) (cluster #2 Of 2), Other |
Other |
8300 |
0.36 |
Functional ≤ 10μM
|
|
Z50523-1-O |
Influenza B Virus (cluster #1 Of 1), Other |
Other |
15 |
0.55 |
Functional ≤ 10μM
|
|
Z50652-1-O |
Influenza A Virus (cluster #1 Of 4), Other |
Other |
6250 |
0.36 |
Functional ≤ 10μM
|
|
Z80249-2-O |
MDCK (Kidney Cells) (cluster #2 Of 2), Other |
Other |
2240 |
0.40 |
ADME/T ≤ 10μM |
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 2), Viral |
Viruses |
5 |
0.58 |
Binding ≤ 10μM
|
|
NRAM-1-V |
Neuraminidase (cluster #1 Of 1), Viral |
Viruses |
4 |
0.59 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.14 |
-2.58 |
-79.64 |
4 |
6 |
0 |
106 |
284.356 |
6 |
↓
|
|
|
|
Analogs
-
4159387
-
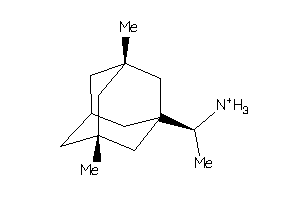
-
4159388
-
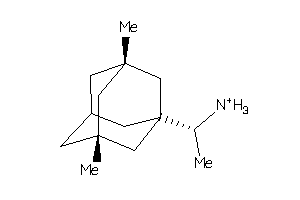
-
40865734
-
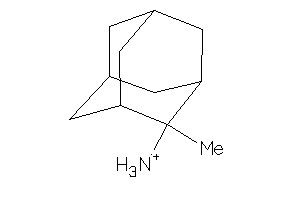
Draw
Identity
99%
90%
80%
70%
Vendors
And 87 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
5.01 |
-42.88 |
3 |
1 |
1 |
28 |
180.315 |
1 |
↓
|
|
|
|
|