|
|
Analogs
-
1657422
-
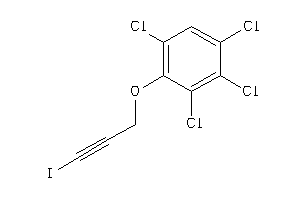
-
32234560
-
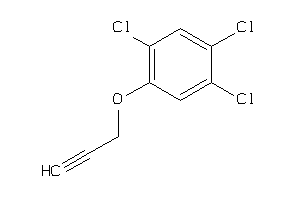
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GALE-1-E |
UDP-glucose 4-epimerase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.63 |
Binding ≤ 10μM
|
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 3), Other |
Other |
300 |
0.65 |
ADME/T ≤ 10μM
|
|
Z80092-1-O |
CHO-K1 (Ovarian Cells) (cluster #1 Of 3), Other |
Other |
600 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.11 |
7.97 |
-4.15 |
0 |
1 |
0 |
9 |
361.393 |
2 |
↓
|
|
|
|
Analogs
-
56544
-
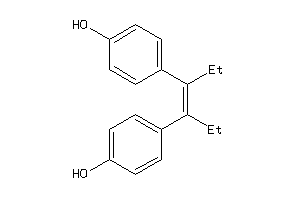
Draw
Identity
99%
90%
80%
70%
Vendors
And 57 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.56 |
Binding ≤ 10μM
|
|
ERR3-1-E |
Estrogen-related Receptor Gamma (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.43 |
Functional ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
ADO-2-E |
Aldehyde Oxidase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.44 |
ADME/T ≤ 10μM
|
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
2500 |
0.39 |
Functional ≤ 10μM
|
|
Z80030-2-O |
ANN-1 (cluster #2 Of 2), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5000 |
0.37 |
Functional ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 3), Other |
Other |
2800 |
0.39 |
ADME/T ≤ 10μM
|
|
Z80092-2-O |
CHO-K1 (Ovarian Cells) (cluster #2 Of 3), Other |
Other |
8500 |
0.35 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
6.1 |
-5.38 |
2 |
2 |
0 |
40 |
268.356 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSTA1-1-E |
Glutathione S-transferase A1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.39 |
Binding ≤ 10μM
|
|
GSTP1-1-E |
Glutathione S-transferase Pi (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.40 |
Binding ≤ 10μM
|
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 3), Other |
Other |
3300 |
0.40 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
8.56 |
-45.9 |
0 |
4 |
-1 |
66 |
302.133 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
13404392
-
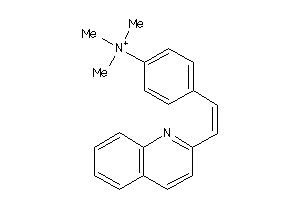
-
5336345
-
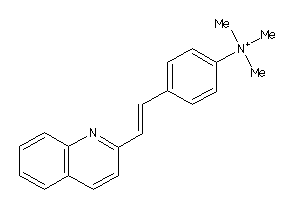
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 6), Other |
Other |
6000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
10.39 |
-8.84 |
0 |
2 |
0 |
16 |
274.367 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3A-1-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE1-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE3-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDE4-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
NMDZ1-5-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
Z50418-3-O |
Trypanosoma Brucei (cluster #3 Of 6), Other |
Other |
1000 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
-2.06 |
-43 |
3 |
1 |
1 |
27 |
194.342 |
3 |
↓
|
|
|
|
Analogs
-
289773
-
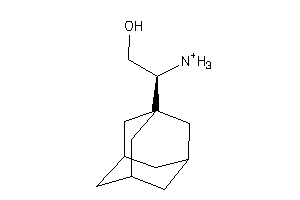
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-3-O |
Trypanosoma Brucei (cluster #3 Of 6), Other |
Other |
9400 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
1.12 |
-37.49 |
4 |
2 |
1 |
48 |
182.287 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.93 |
0.91 |
-2.01 |
3 |
2 |
0 |
46 |
181.279 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.36 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.36 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.32 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10 |
0.36 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.36 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
119 |
0.31 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
5100 |
0.24 |
Functional ≤ 10μM
|
|
Z80244-2-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #2 Of 7), Other |
Other |
6800 |
0.23 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
1700 |
0.26 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
5.59 |
-18.74 |
3 |
8 |
0 |
117 |
449.517 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.26 |
5.12 |
-53.22 |
2 |
8 |
-1 |
119 |
448.509 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.26 |
5.03 |
-53.98 |
2 |
8 |
-1 |
119 |
448.509 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
8000 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.13 |
-2.23 |
-13.05 |
0 |
7 |
0 |
97 |
205.195 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
1000 |
0.44 |
Functional ≤ 10μM
|
|
Z50725-2-O |
Trypanosoma Brucei Rhodesiense (cluster #2 Of 7), Other |
Other |
700 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
1.08 |
-9.3 |
0 |
6 |
0 |
72 |
279.321 |
5 |
↓
|
|
|
|
Analogs
-
367100
-
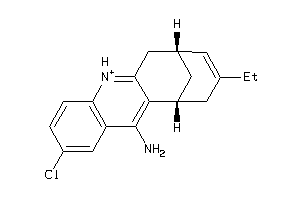
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
257 |
0.44 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
762 |
0.41 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
2150 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.77 |
9.05 |
-26.42 |
3 |
2 |
1 |
40 |
299.825 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.77 |
8.68 |
-6.24 |
2 |
2 |
0 |
39 |
298.817 |
1 |
↓
|
|
|
|
Analogs
-
367097
-
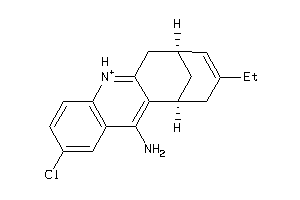
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
257 |
0.44 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
762 |
0.41 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
2150 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.77 |
9.06 |
-26.56 |
3 |
2 |
1 |
40 |
299.825 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.77 |
8.69 |
-6.12 |
2 |
2 |
0 |
39 |
298.817 |
1 |
↓
|
|
|
|
Analogs
-
3978828
-
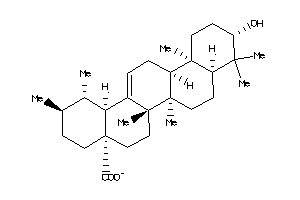
-
3978829
-
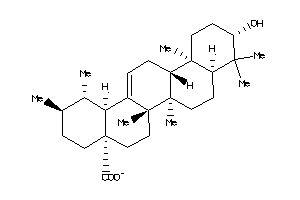
-
4273370
-
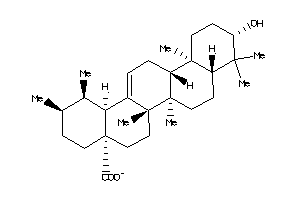
-
4273371
-
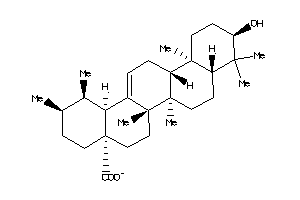
-
4273372
-
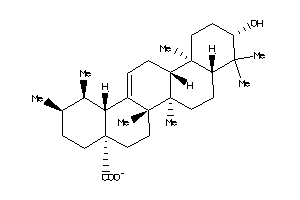
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHI1-2-E |
11-beta-hydroxysteroid Dehydrogenase 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.24 |
Binding ≤ 10μM
|
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.22 |
Binding ≤ 10μM
|
|
PA21B-3-E |
Phospholipase A2 Group 1B (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.23 |
Binding ≤ 10μM
|
|
PA2A-1-E |
Phospholipase A2 Isozyme PLA-A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GA-3-E |
Phospholipase A2, Membrane Associated (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GD-2-E |
Group IID Secretory Phospholipase A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
PA2GE-2-E |
Group IIE Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PA2GF-2-E |
Group IIF Secretory Phospholipase A2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-2-E |
T-cell Protein-tyrosine Phosphatase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6700 |
0.22 |
Binding ≤ 10μM |
|
PYGM-1-E |
Muscle Glycogen Phosphorylase (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.21 |
Binding ≤ 10μM |
|
Q7T3S7-1-E |
Phospholipase A2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.24 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1430 |
0.25 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-4-O |
Trypanosoma Brucei (cluster #4 Of 6), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50420-3-O |
Trypanosoma Brucei Brucei (cluster #3 Of 7), Other |
Other |
2200 |
0.24 |
Functional ≤ 10μM
|
|
Z50466-5-O |
Trypanosoma Cruzi (cluster #5 Of 8), Other |
Other |
4000 |
0.23 |
Functional ≤ 10μM
|
|
Z50472-2-O |
Toxoplasma Gondii (cluster #2 Of 4), Other |
Other |
1000 |
0.25 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
1800 |
0.24 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
4400 |
0.23 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
12.05 |
-49.78 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.79 |
10.29 |
-5.45 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
1650 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.41 |
0.12 |
-9.41 |
1 |
2 |
0 |
29 |
322.235 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
4.29 |
-16.48 |
2 |
9 |
0 |
138 |
252.186 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.29 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
960 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
1.74 |
-20.31 |
1 |
7 |
0 |
101 |
370.405 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.40 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
1010 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
8.37 |
-16.62 |
1 |
5 |
0 |
75 |
270.288 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
150 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.19 |
-16.76 |
1 |
5 |
0 |
74 |
298.342 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.37 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
820 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.29 |
8.44 |
-16.71 |
1 |
5 |
0 |
75 |
288.278 |
6 |
↓
|
|
|
|
Analogs
-
36179887
-
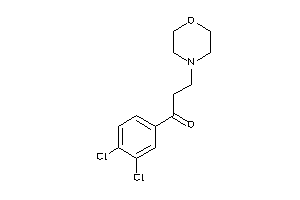
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
980 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
4.75 |
-7.24 |
0 |
3 |
0 |
30 |
253.729 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-3-O |
Trypanosoma Brucei (cluster #3 Of 6), Other |
Other |
5280 |
0.49 |
Functional ≤ 10μM |
|
Z50652-2-O |
Influenza A Virus (cluster #2 Of 4), Other |
Other |
160 |
0.63 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
6.43 |
-42.28 |
2 |
1 |
1 |
17 |
206.353 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.60 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.60 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.60 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
55 |
0.56 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
35 |
0.58 |
Binding ≤ 10μM
|
|
CDK4-2-E |
Cyclin-dependent Kinase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.51 |
Binding ≤ 10μM
|
|
CDK5-1-E |
Cyclin-dependent Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.56 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Binding ≤ 10μM |
|
GSK3A-2-E |
Glycogen Synthase Kinase-3 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.50 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
350 |
0.50 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.45 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.54 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
180 |
0.52 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
5.1 |
-16.01 |
2 |
5 |
0 |
71 |
241.25 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.39 |
4.84 |
-40.96 |
3 |
5 |
1 |
72 |
242.258 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
150 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
9.13 |
-22.54 |
2 |
7 |
0 |
115 |
346.346 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNA1-1-E |
Cyclin A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
380 |
0.36 |
Binding ≤ 10μM
|
|
CCNA2-1-E |
Cyclin A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
380 |
0.36 |
Binding ≤ 10μM
|
|
CCNB1-2-E |
G2/mitotic-specific Cyclin B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CCNB2-2-E |
G2/mitotic-specific Cyclin B2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CCNB3-2-E |
G2/mitotic-specific Cyclin B3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.34 |
Binding ≤ 10μM
|
|
CDK2-2-E |
Cyclin-dependent Kinase 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
380 |
0.36 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.40 |
Binding ≤ 10μM
|
|
GSK3B-1-E |
Glycogen Synthase Kinase-3 Beta (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
90 |
0.39 |
Binding ≤ 10μM |
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
740 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
3.25 |
-14.88 |
3 |
8 |
0 |
128 |
359.725 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.66 |
4.01 |
-47.82 |
2 |
8 |
-1 |
131 |
358.717 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.90 |
4.58 |
-93.91 |
7 |
8 |
2 |
126 |
405.506 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
-5.91 |
-77.64 |
9 |
7 |
2 |
139 |
283.339 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Dihydrofolate Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Functional ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
10 |
0.66 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
7000 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
80 |
0.58 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
9900 |
0.41 |
Functional ≤ 10μM |
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
700 |
0.51 |
Functional ≤ 10μM
|
|
Z80186-3-O |
K562 (Erythroleukemia Cells) (cluster #3 Of 3), Other |
Other |
5000 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
6.53 |
-28.58 |
5 |
4 |
1 |
79 |
249.725 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.84 |
6.08 |
-5.33 |
4 |
4 |
0 |
78 |
248.717 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7800 |
0.36 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
950 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
-2.04 |
-50.03 |
1 |
4 |
1 |
55 |
296.412 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
3300 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.20 |
2.41 |
-35.27 |
2 |
2 |
1 |
24 |
318.525 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.46 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
550 |
0.52 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.63 |
-1.36 |
-51.18 |
1 |
4 |
1 |
55 |
256.347 |
5 |
↓
|
|
|
|
Analogs
-
4159387
-
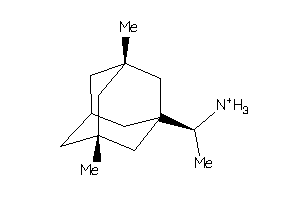
-
4159388
-
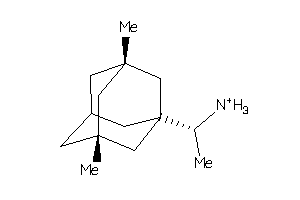
-
40865734
-
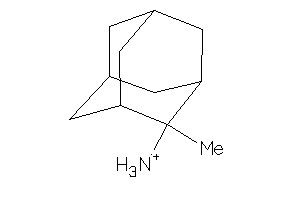
Draw
Identity
99%
90%
80%
70%
Vendors
And 87 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
5.01 |
-42.88 |
3 |
1 |
1 |
28 |
180.315 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
34543218
-
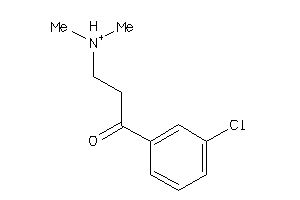
-
39372435
-
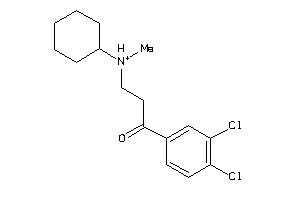
-
1572430
-
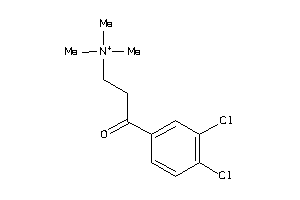
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.51 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
1000 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
1.99 |
-46.01 |
1 |
2 |
1 |
21 |
247.145 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
340 |
0.57 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
680 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.70 |
7.73 |
-54.13 |
1 |
5 |
1 |
67 |
223.252 |
5 |
↓
|
|
|
|
Analogs
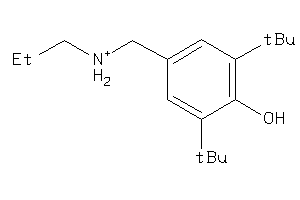
-
22141174
-
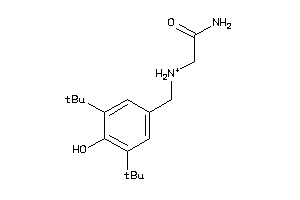
-
1533450
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TYTR-1-E |
Trypanothione Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
3010 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
-1.51 |
-38.79 |
4 |
3 |
1 |
57 |
280.432 |
6 |
↓
|
|
|
|
Analogs
-
6535287
-
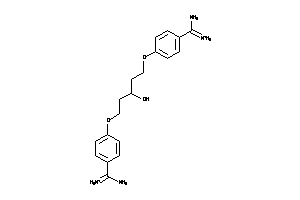
-
6535291
-
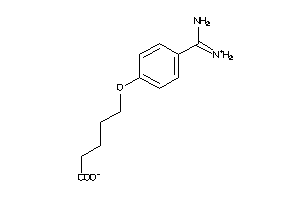
-
6535292
-
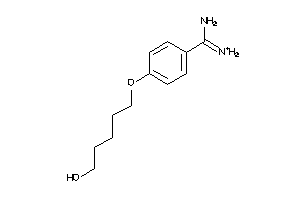
-
34928597
-
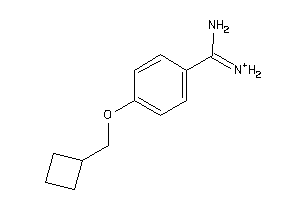
Draw
Identity
99%
90%
80%
70%
Vendors
And 50 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S100B-2-E |
S-100 Protein Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.34 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1160 |
0.33 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2300 |
0.32 |
Binding ≤ 10μM
|
|
Z104302-2-O |
Glutamate NMDA Receptor (cluster #2 Of 7), Other |
Other |
3060 |
0.31 |
Binding ≤ 10μM |
|
Z101865-1-O |
Leishmania Chagasi (cluster #1 Of 1), Other |
Other |
1110 |
0.33 |
Functional ≤ 10μM
|
|
Z101870-2-O |
Leishmania Guyanensis (cluster #2 Of 2), Other |
Other |
900 |
0.34 |
Functional ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
84 |
0.40 |
Functional ≤ 10μM
|
|
Z50380-1-O |
Mycobacterium Smegmatis (cluster #1 Of 4), Other |
Other |
9100 |
0.28 |
Functional ≤ 10μM
|
|
Z50418-5-O |
Trypanosoma Brucei (cluster #5 Of 6), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
|
Z50420-1-O |
Trypanosoma Brucei Brucei (cluster #1 Of 7), Other |
Other |
3 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-9-O |
Plasmodium Falciparum (cluster #9 Of 22), Other |
Other |
251 |
0.37 |
Functional ≤ 10μM
|
|
Z50426-1-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #1 Of 9), Other |
Other |
6 |
0.46 |
Functional ≤ 10μM
|
|
Z50457-1-O |
Leishmania Amazonensis (cluster #1 Of 3), Other |
Other |
460 |
0.35 |
Functional ≤ 10μM
|
|
Z50458-4-O |
Leishmania Braziliensis (cluster #4 Of 4), Other |
Other |
9800 |
0.28 |
Functional ≤ 10μM
|
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
9800 |
0.28 |
Functional ≤ 10μM
|
|
Z50460-1-O |
Leishmania Major (cluster #1 Of 4), Other |
Other |
7500 |
0.29 |
Functional ≤ 10μM
|
|
Z50461-1-O |
Leishmania Mexicana (cluster #1 Of 3), Other |
Other |
9568 |
0.28 |
Functional ≤ 10μM
|
|
Z50466-4-O |
Trypanosoma Cruzi (cluster #4 Of 8), Other |
Other |
7100 |
0.29 |
Functional ≤ 10μM
|
|
Z50467-2-O |
Trichomonas Vaginalis (cluster #2 Of 3), Other |
Other |
3815 |
0.30 |
Functional ≤ 10μM
|
|
Z50468-4-O |
Giardia Intestinalis (cluster #4 Of 4), Other |
Other |
4079 |
0.30 |
Functional ≤ 10μM
|
|
Z50473-4-O |
Plasmodium Berghei (cluster #4 Of 5), Other |
Other |
2942 |
0.31 |
Functional ≤ 10μM
|
|
Z50594-6-O |
Mus Musculus (cluster #6 Of 9), Other |
Other |
3 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
2500 |
0.31 |
Functional ≤ 10μM
|
|
Z50725-1-O |
Trypanosoma Brucei Rhodesiense (cluster #1 Of 7), Other |
Other |
50 |
0.41 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
40 |
0.41 |
Functional ≤ 10μM
|
|
Z81135-4-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #4 Of 4), Other |
Other |
2100 |
0.32 |
Functional ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 2), Other |
Other |
1030 |
0.34 |
ADME/T ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
2700 |
0.31 |
ADME/T ≤ 10μM
|
|
Z81135-2-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #2 Of 6), Other |
Other |
6000 |
0.29 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.49 |
5.45 |
-74.02 |
8 |
6 |
2 |
122 |
342.443 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.68 |
1.99 |
-2.42 |
1 |
1 |
0 |
20 |
114.188 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-6-O |
Trypanosoma Brucei (cluster #6 Of 6), Other |
Other |
5400 |
0.49 |
Functional ≤ 10μM
|
|
Z50725-4-O |
Trypanosoma Brucei Rhodesiense (cluster #4 Of 7), Other |
Other |
5400 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.50 |
-3.35 |
-173.27 |
3 |
8 |
-2 |
148 |
261.107 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.50 |
-4.49 |
-96.88 |
4 |
8 |
-1 |
145 |
262.115 |
5 |
↓
|
|
|
|
Analogs
-
4802968
-
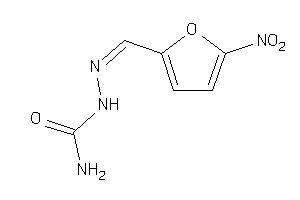
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
-0.73 |
-20.5 |
3 |
8 |
0 |
126 |
198.138 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50418-1-O |
Trypanosoma Brucei (cluster #1 Of 6), Other |
Other |
3000 |
0.43 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
755 |
0.48 |
Functional ≤ 10μM |
|
Z50426-9-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #9 Of 9), Other |
Other |
440 |
0.49 |
Functional ≤ 10μM
|
|
Z50459-8-O |
Leishmania Donovani (cluster #8 Of 8), Other |
Other |
2680 |
0.43 |
Functional ≤ 10μM
|
|
Z50466-2-O |
Trypanosoma Cruzi (cluster #2 Of 8), Other |
Other |
220 |
0.52 |
Functional ≤ 10μM
|
|
Z50725-7-O |
Trypanosoma Brucei Rhodesiense (cluster #7 Of 7), Other |
Other |
600 |
0.48 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1500 |
0.45 |
Functional ≤ 10μM
|
|
Z80695-2-O |
BE (Colon Adenocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
1280 |
0.46 |
Functional ≤ 10μM
|
|
Z81240-1-O |
MAC15A (Colon Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
3950 |
0.42 |
Functional ≤ 10μM
|
|
Z80583-3-O |
Vero (Kidney Cells) (cluster #3 Of 3), Other |
Other |
1050 |
0.47 |
ADME/T ≤ 10μM |
|
Z81057-4-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #4 Of 5), Other |
Other |
1180 |
0.46 |
ADME/T ≤ 10μM
|
|
Z81135-5-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #5 Of 6), Other |
Other |
1120 |
0.46 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
0.21 |
-16.41 |
0 |
2 |
0 |
17 |
232.286 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FPPS-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
82 |
0.58 |
Binding ≤ 10μM
|
|
Q0GKD7-1-E |
Farnesyl Pyrophosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
|
Q197X6-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.59 |
Binding ≤ 10μM
|
|
Q8WS26-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Binding ≤ 10μM
|
|
Z50418-6-O |
Trypanosoma Brucei (cluster #6 Of 6), Other |
Other |
8600 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-17-O |
Plasmodium Falciparum (cluster #17 Of 22), Other |
Other |
1400 |
0.48 |
Functional ≤ 10μM
|
|
Z50459-5-O |
Leishmania Donovani (cluster #5 Of 8), Other |
Other |
2300 |
0.46 |
Functional ≤ 10μM
|
|
Z50472-3-O |
Toxoplasma Gondii (cluster #3 Of 4), Other |
Other |
490 |
0.52 |
Functional ≤ 10μM
|
|
Z50594-2-O |
Mus Musculus (cluster #2 Of 9), Other |
Other |
300 |
0.54 |
Functional ≤ 10μM
|
|
Z50725-4-O |
Trypanosoma Brucei Rhodesiense (cluster #4 Of 7), Other |
Other |
8600 |
0.42 |
Functional ≤ 10μM
|
|
Z50759-2-O |
Dictyostelium Discoideum (cluster #2 Of 2), Other |
Other |
3000 |
0.45 |
Functional ≤ 10μM
|
|
Z80954-2-O |
HFF (Foreskin Fibroblasts) (cluster #2 Of 4), Other |
Other |
2400 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.23 |
-3.55 |
-231.08 |
2 |
8 |
-3 |
157 |
280.089 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.23 |
-2.31 |
-381.96 |
1 |
8 |
-4 |
159 |
279.081 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.23 |
-4.53 |
-107.38 |
3 |
8 |
-2 |
154 |
281.097 |
4 |
↓
|
|