|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50501-1-O |
Gallus Gallus (cluster #1 Of 3), Other |
Other |
7300 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
-0.6 |
-9.42 |
2 |
4 |
0 |
56 |
322.799 |
1 |
↓
|
|
|
|
Analogs
-
3823565
-
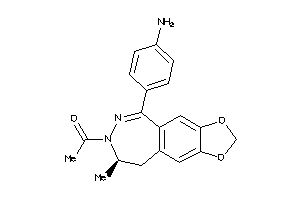
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-0.46 |
-10.92 |
2 |
6 |
0 |
77 |
337.379 |
1 |
↓
|
|
|
|
Analogs
-
2121
-
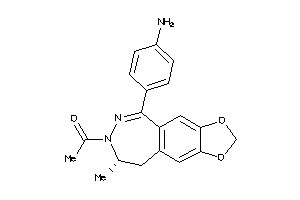
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-0.49 |
-10.37 |
2 |
6 |
0 |
77 |
337.379 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
9000 |
0.32 |
Binding ≤ 10μM
|
|
GRIA1-1-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.32 |
Functional ≤ 10μM
|
|
Z50501-1-O |
Gallus Gallus (cluster #1 Of 3), Other |
Other |
9500 |
0.32 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
9900 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
5.48 |
-12.21 |
2 |
5 |
0 |
69 |
293.326 |
1 |
↓
|
|
|
|
Analogs
-
3834201
-
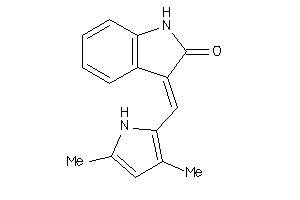
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.46 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
84 |
0.55 |
Binding ≤ 10μM
|
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7080 |
0.40 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
160 |
0.53 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
660 |
0.48 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.48 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.53 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1040 |
0.47 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
884 |
0.47 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.57 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4050 |
0.42 |
Functional ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4540 |
0.42 |
Functional ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.58 |
Functional ≤ 10μM
|
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
5 |
0.65 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
85 |
0.55 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.40 |
Functional ≤ 10μM
|
|
Z50501-1-O |
Gallus Gallus (cluster #1 Of 3), Other |
Other |
85 |
0.55 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
4540 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
6.69 |
-8.41 |
2 |
3 |
0 |
49 |
238.29 |
1 |
↓
|
|
|
|
Analogs
-
1997624
-
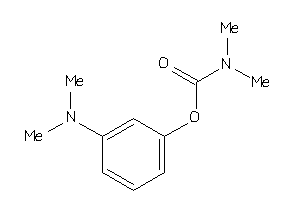
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.34 |
7.92 |
-35.58 |
0 |
4 |
1 |
30 |
223.296 |
3 |
↓
|
|