|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITPR1-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM |
|
ITPR2-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.39 |
Binding ≤ 10μM |
|
ITPR3-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.48 |
Binding ≤ 10μM
|
|
ITPR1-1-E |
Inositol 1,4,5-trisphosphate Receptor Type 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.44 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 3), Other |
Other |
10 |
0.47 |
Binding ≤ 10μM |
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 12), Other |
Other |
90 |
0.41 |
Functional ≤ 10μM
|
|
Z50597-4-O |
Rattus Norvegicus (cluster #4 Of 12), Other |
Other |
119 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.71 |
-8.52 |
-463.16 |
4 |
15 |
-5 |
275 |
415.053 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.71 |
-8.49 |
-497.6 |
4 |
15 |
-5 |
275 |
415.053 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.71 |
-7.37 |
-637.17 |
3 |
15 |
-6 |
278 |
414.045 |
6 |
↓
|
|
|
|
Analogs
-
39293733
-
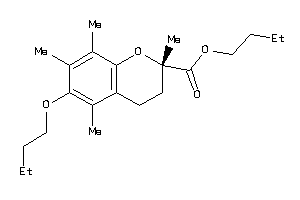
-
39293735
-
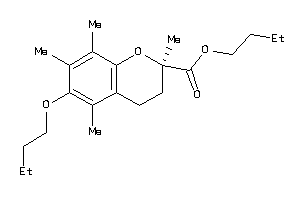
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
5.96 |
-49.86 |
1 |
4 |
-1 |
70 |
249.286 |
1 |
↓
|
|
|
|
Analogs
-
39293733
-

-
39293735
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
5.98 |
-50.1 |
1 |
4 |
-1 |
70 |
249.286 |
1 |
↓
|
|
|
|
Analogs
-
4098885
-
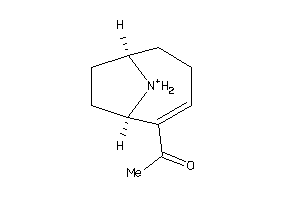
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
90 |
0.82 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB-3-E |
Acetylcholine Receptor Protein Beta Chain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
19 |
0.90 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.94 |
Binding ≤ 10μM
|
|
ACHG-3-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.81 |
Functional ≤ 10μM
|
|
Z104287-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 3), Other |
Other |
25 |
0.89 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
1 |
1.05 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
48 |
0.85 |
Binding ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
4.05 |
-42.61 |
2 |
2 |
1 |
34 |
166.244 |
1 |
↓
|
|
|
|
Analogs
-
4098884
-
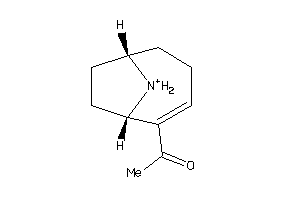
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA2-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA5-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA6-3-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA7-6-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHA9-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB-3-E |
Acetylcholine Receptor Protein Beta Chain (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB2-5-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB3-3-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHB4-2-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
5 |
0.97 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.94 |
Binding ≤ 10μM
|
|
ACHG-3-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.05 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.81 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
48 |
0.85 |
Binding ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
-0.69 |
-42.22 |
2 |
2 |
1 |
33 |
166.244 |
1 |
↓
|
|
|
|
Analogs
-
600699
-
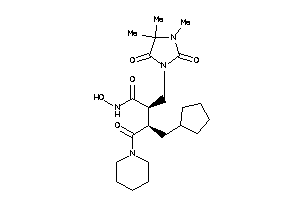
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP13-1-E |
Matrix Metalloproteinase 13 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
527 |
0.28 |
Binding ≤ 10μM |
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.37 |
Binding ≤ 10μM |
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
100 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.07 |
-1.05 |
-17.22 |
2 |
9 |
0 |
110 |
436.553 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.07 |
-0.49 |
-66.15 |
1 |
9 |
-1 |
113 |
435.545 |
7 |
↓
|
|
|
|
Analogs
-
899213
-
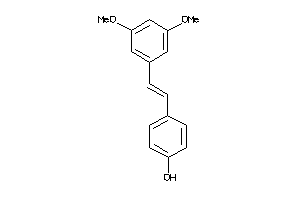
-
13456778
-
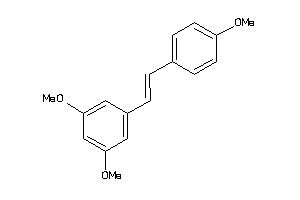
-
3225
-
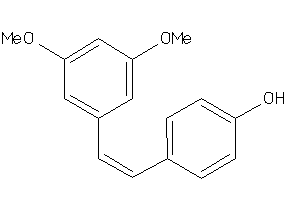
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
75 |
0.50 |
Binding ≤ 10μM
|
|
ESR1-1-E |
Estrogen Receptor Alpha (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
132 |
0.48 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.40 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.40 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.40 |
Functional ≤ 10μM
|
|
TBB3-3-E |
Tubulin Beta-3 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Functional ≤ 10μM
|
|
TBB4A-3-E |
Tubulin Beta-4 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Functional ≤ 10μM
|
|
TBB4B-3-E |
Tubulin Beta-2 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Functional ≤ 10μM
|
|
TBB8-3-E |
Tubulin Beta-8 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.38 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
3800 |
0.38 |
Functional ≤ 10μM
|
|
Z80054-4-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
80 |
0.50 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
240 |
0.46 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
54 |
0.51 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
120 |
0.48 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
218 |
0.47 |
Functional ≤ 10μM
|
|
Z80526-2-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #2 Of 6), Other |
Other |
300 |
0.46 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
220 |
0.47 |
Functional ≤ 10μM
|
|
Z81001-1-O |
TSGH 9201 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
324 |
0.45 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
253 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
280 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
2.19 |
-7.53 |
0 |
3 |
0 |
27 |
270.328 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
5744533
-
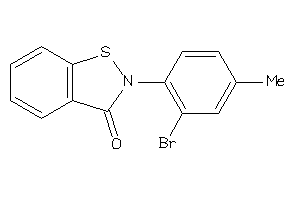
-
6583578
-
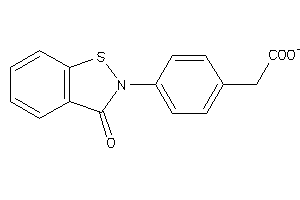
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPI-1-E |
Mannose-6-phosphate Isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6400 |
0.45 |
Binding ≤ 10μM
|
|
Z50591-1-O |
Bos Taurus (cluster #1 Of 2), Other |
Other |
3000 |
0.48 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
8.02 |
-12.19 |
0 |
2 |
0 |
22 |
227.288 |
1 |
↓
|
|
|
|
Analogs
-
1591265
-
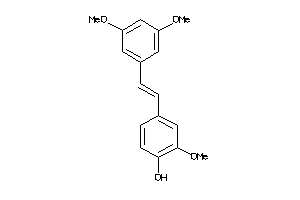
-
1591410
-
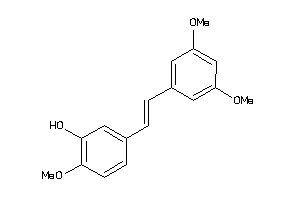
-
1611146
-
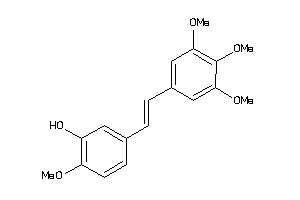
-
1612318
-
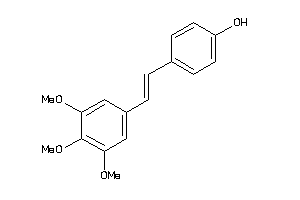
-
1616881
-
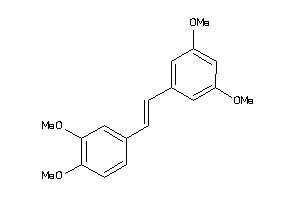
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.37 |
Binding ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.37 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.43 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.37 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.37 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.36 |
Functional ≤ 10μM
|
|
TBB3-3-E |
Tubulin Beta-3 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Functional ≤ 10μM
|
|
TBB4A-3-E |
Tubulin Beta-4 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Functional ≤ 10μM
|
|
TBB4B-3-E |
Tubulin Beta-2 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Functional ≤ 10μM
|
|
TBB8-3-E |
Tubulin Beta-8 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2500 |
0.36 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
13 |
0.50 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
15 |
0.50 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
17 |
0.49 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z81001-1-O |
TSGH 9201 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
60 |
0.46 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
16 |
0.50 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
9 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.18 |
2.94 |
-9.07 |
0 |
4 |
0 |
36 |
300.354 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50591-1-O |
Bos Taurus (cluster #1 Of 2), Other |
Other |
10000 |
0.22 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.09 |
13.62 |
-23.86 |
2 |
4 |
0 |
58 |
456.592 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LEF-1-B |
Anthrax Lethal Factor (cluster #1 Of 2), Bacterial |
Bacteria |
1200 |
0.31 |
Binding ≤ 10μM
|
|
ADA10-1-E |
ADAM10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.31 |
Binding ≤ 10μM
|
|
ADA17-2-E |
ADAM17 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.38 |
Binding ≤ 10μM
|
|
BMP1-1-E |
Bone Morphogenetic Protein 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
8600 |
0.26 |
Binding ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 11), Eukaryotic |
Eukaryotes |
7800 |
0.26 |
Binding ≤ 10μM
|
|
MMP1-1-E |
Matrix Metalloproteinase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
96 |
0.36 |
Binding ≤ 10μM
|
|
MMP12-1-E |
Macrophage Metalloelastase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
MMP13-3-E |
Matrix Metalloproteinase 13 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM
|
|
MMP14-2-E |
Matrix Metalloproteinase 14 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.42 |
Binding ≤ 10μM
|
|
MMP16-1-E |
Matrix Metalloproteinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.41 |
Binding ≤ 10μM
|
|
MMP25-1-E |
Matrix Metalloproteinase-25 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.45 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
|
MMP7-1-E |
Matrix Metalloproteinase 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
736 |
0.32 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.42 |
Binding ≤ 10μM |
|
MMP9-1-E |
Matrix Metalloproteinase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.41 |
Binding ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
50 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.65 |
-24.43 |
2 |
8 |
0 |
109 |
393.465 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.55 |
1.28 |
-51.2 |
1 |
8 |
-1 |
115 |
392.457 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.37 |
3.89 |
-61.96 |
1 |
8 |
-1 |
112 |
392.457 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.33 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.33 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.40 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.40 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
1000 |
0.37 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
12 |
0.48 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80792-1-O |
Colon 26 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
269 |
0.40 |
Functional ≤ 10μM
|
|
Z81001-1-O |
TSGH 9201 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
1 |
0.55 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
45 |
0.45 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
45 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
1.02 |
-9.26 |
2 |
5 |
0 |
62 |
315.369 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Binding ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.35 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.34 |
Binding ≤ 10μM |
|
Q25270-2-E |
Beta Tubulin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Functional ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Functional ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4920 |
0.32 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
530 |
0.38 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.38 |
Functional ≤ 10μM
|
|
TBB3-3-E |
Tubulin Beta-3 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB4A-3-E |
Tubulin Beta-4 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB4B-3-E |
Tubulin Beta-2 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
TBB8-3-E |
Tubulin Beta-8 Chain (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Functional ≤ 10μM
|
|
Z100495-1-O |
BMEC (Brain Microvessel Endothelial Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z100742-1-O |
MeWo (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM |
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
188 |
0.41 |
Functional ≤ 10μM |
|
Z103208-1-O |
AU565 (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z103407-2-O |
SW1990 (cluster #2 Of 2), Other |
Other |
250 |
0.40 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
2000 |
0.35 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
18 |
0.47 |
Functional ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80013-2-O |
A10 (cluster #2 Of 2), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80035-5-O |
B16 (Melanoma Cells) (cluster #5 Of 7), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80064-4-O |
CCRF-CEM (T-cell Leukemia) (cluster #4 Of 9), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80071-2-O |
CEM-0 (T-cell Leukemia) (cluster #2 Of 2), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80099-3-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
2756 |
0.34 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
36 |
0.45 |
Functional ≤ 10μM
|
|
Z80121-1-O |
DMS-79 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
124 |
0.42 |
Functional ≤ 10μM
|
|
Z80125-9-O |
DU-145 (Prostate Carcinoma) (cluster #9 Of 9), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80136-2-O |
FM3A (Breast Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
42 |
0.45 |
Functional ≤ 10μM |
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
94 |
0.43 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM |
|
Z80193-1-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #1 Of 12), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80211-2-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80229-1-O |
MCF7-DOX (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80254-1-O |
MES-SA (Uterine Sarcoma Cells) (cluster #1 Of 2), Other |
Other |
13 |
0.48 |
Functional ≤ 10μM
|
|
Z80255-2-O |
MES-SA/DxS (Uterine Sarcoma Cells) (cluster #2 Of 2), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
82 |
0.43 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80285-2-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #2 Of 4), Other |
Other |
16 |
0.47 |
Functional ≤ 10μM
|
|
Z80353-1-O |
OVCAR (cluster #1 Of 1), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
5400 |
0.32 |
Functional ≤ 10μM
|
|
Z80471-3-O |
SH-SY5 (Bone Marrow Neuroblastoma Cells) (cluster #3 Of 3), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z80485-3-O |
SK-MEL-28 (Melanoma Cells) (cluster #3 Of 6), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM |
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
45 |
0.45 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
8 |
0.49 |
Functional ≤ 10μM
|
|
Z80697-3-O |
Bel-7402 (Hepatoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z80711-1-O |
NCI/ADR-RES (Breast Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80792-1-O |
Colon 26 (Colon Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
9 |
0.49 |
Functional ≤ 10μM
|
|
Z80798-1-O |
CPT30 Cell Line (cluster #1 Of 1), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z80847-3-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80852-2-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM
|
|
Z80874-2-O |
CEM (T-cell Leukemia) (cluster #2 Of 7), Other |
Other |
12 |
0.48 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z80900-1-O |
HA22T Cell Line (cluster #1 Of 1), Other |
Other |
2708 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-6-O |
HCT-116 (Colon Carcinoma Cells) (cluster #6 Of 9), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z80967-2-O |
Hone-1 Cell Line (cluster #2 Of 2), Other |
Other |
7 |
0.50 |
Functional ≤ 10μM
|
|
Z81001-1-O |
TSGH 9201 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
38 |
0.45 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
6 |
0.50 |
Functional ≤ 10μM
|
|
Z81024-5-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #5 Of 8), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
2 |
0.53 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM
|
|
Z81065-3-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
5 |
0.51 |
Functional ≤ 10μM |
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z81233-2-O |
M21 (Melanoma Cells) (cluster #2 Of 2), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
4 |
0.51 |
Functional ≤ 10μM |
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
43 |
0.45 |
Functional ≤ 10μM |
|
Z81331-6-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #6 Of 6), Other |
Other |
23 |
0.46 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3 |
0.52 |
Functional ≤ 10μM
|
|
Z100495-1-O |
BMEC (Brain Microvessel Endothelial Cells) (cluster #1 Of 1), Other |
Other |
4 |
0.51 |
ADME/T ≤ 10μM
|
|
Z80013-2-O |
A10 (cluster #2 Of 2), Other |
Other |
23 |
0.46 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.52 |
ADME/T ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 4), Other |
Other |
2 |
0.53 |
ADME/T ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
ADME/T ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 5), Other |
Other |
5 |
0.51 |
ADME/T ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
3 |
0.52 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
5.32 |
-10.67 |
1 |
5 |
0 |
57 |
316.353 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AT2A1-1-E |
Sarcoplasmic/endoplasmic Reticulum Calcium ATP-ase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
AT2A2-1-E |
Sarcoplasmic/endoplasmic Reticulum Calcium ATPase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.24 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3981 |
0.16 |
Functional ≤ 10μM
|
|
Z50591-2-O |
Bos Taurus (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.99 |
3.5 |
-16.81 |
2 |
12 |
0 |
171 |
650.762 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MPI-1-E |
Mannose-6-phosphate Isomerase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.47 |
Binding ≤ 10μM
|
|
Z50591-1-O |
Bos Taurus (cluster #1 Of 2), Other |
Other |
3600 |
0.45 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
8.53 |
-9.99 |
0 |
2 |
0 |
22 |
261.733 |
1 |
↓
|
|