|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.76 |
-4.94 |
-15.02 |
2 |
6 |
0 |
84 |
220.612 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
39908017
-
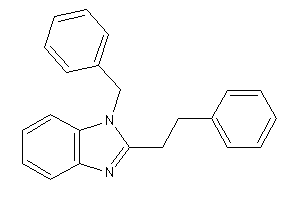
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
410 |
0.37 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
13.15 |
-9.89 |
0 |
3 |
0 |
23 |
315.42 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.40 |
13.62 |
-31.98 |
1 |
3 |
1 |
24 |
316.428 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
188 |
0.35 |
Binding ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
188 |
0.35 |
Binding ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
188 |
0.35 |
Binding ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
188 |
0.35 |
Binding ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.77 |
-0.75 |
-361.17 |
2 |
15 |
-4 |
241 |
465.166 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.77 |
-1.91 |
-224.76 |
3 |
15 |
-3 |
238 |
466.174 |
8 |
↓
|
|
|
|
Analogs
-
37847064
-
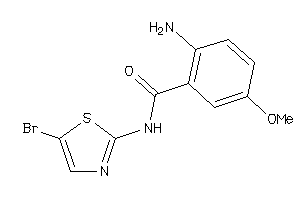
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-2-O |
Hepatitis B Virus (cluster #2 Of 3), Other |
Other |
7600 |
0.42 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.50 |
4.99 |
-13.27 |
2 |
4 |
0 |
62 |
313.176 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.50 |
5.99 |
-55.3 |
1 |
4 |
-1 |
65 |
312.168 |
2 |
↓
|
|
|
|
Analogs
-
37847064
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-2-O |
Hepatitis B Virus (cluster #2 Of 3), Other |
Other |
2900 |
0.48 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
4.28 |
-13.53 |
2 |
4 |
0 |
62 |
299.149 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.30 |
5.28 |
-56.76 |
1 |
4 |
-1 |
65 |
298.141 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.51 |
-3.18 |
-12.38 |
2 |
6 |
0 |
84 |
242.206 |
2 |
↓
|
|
|
|
Analogs
-
4400
-
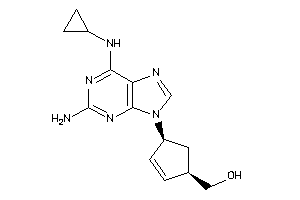
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.22 |
5.36 |
-10.23 |
4 |
7 |
0 |
102 |
286.339 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.22 |
5.75 |
-28.77 |
5 |
7 |
1 |
103 |
287.347 |
4 |
↓
|
|
|
|
Analogs
-
5378904
-
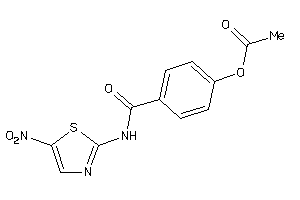
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
7.98 |
-18.51 |
1 |
8 |
0 |
114 |
307.287 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.54 |
6.55 |
-43.57 |
0 |
8 |
-1 |
120 |
306.279 |
5 |
↓
|
|
|
|
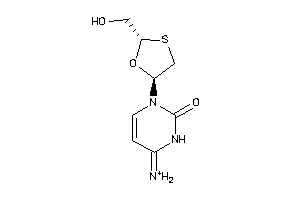
Analogs
-
16952920
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z100081-1-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #1 Of 4), Other |
Other |
27 |
0.71 |
Functional ≤ 10μM
|
|
Z101829-1-O |
HBV Genotype D (cluster #1 Of 1), Other |
Other |
460 |
0.59 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
400 |
0.60 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
2200 |
0.53 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
81 |
0.66 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
969 |
0.56 |
Functional ≤ 10μM
|
|
Z50658-1-O |
Human Immunodeficiency Virus 2 (cluster #1 Of 4), Other |
Other |
2000 |
0.53 |
Functional ≤ 10μM
|
|
Z50677-1-O |
Human Immunodeficiency Virus (cluster #1 Of 3), Other |
Other |
60 |
0.67 |
Functional ≤ 10μM
|
|
Z50681-1-O |
Duck Hepatitis B Virus (cluster #1 Of 2), Other |
Other |
44 |
0.69 |
Functional ≤ 10μM
|
|
Z50755-1-O |
Anas Sp. (cluster #1 Of 1), Other |
Other |
100 |
0.65 |
Functional ≤ 10μM |
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
7000 |
0.48 |
Functional ≤ 10μM |
|
Z80947-1-O |
Hepatoblastoma Cell Line (cluster #1 Of 1), Other |
Other |
70 |
0.67 |
Functional ≤ 10μM
|
|
Z80949-1-O |
Hepatoma Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.75 |
Functional ≤ 10μM |
|
Z81020-1-O |
HepG2 (Hepatoblastoma Cells) (cluster #1 Of 8), Other |
Other |
8 |
0.76 |
Functional ≤ 10μM
|
|
Q72547-1-V |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase (cluster #1 Of 6), Viral |
Viruses |
42 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.09 |
-2.17 |
-16.53 |
3 |
6 |
0 |
90 |
229.261 |
2 |
↓
|
|
|
|
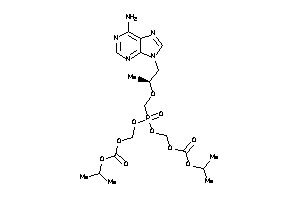
Analogs
-
11616672
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
9.77 |
-22.6 |
2 |
15 |
0 |
185 |
519.448 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50527-1-O |
Human Herpesvirus 3 (cluster #1 Of 3), Other |
Other |
140 |
0.53 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
8650 |
0.39 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
700 |
0.48 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
8100 |
0.40 |
Functional ≤ 10μM |
|
Z81020-5-O |
HepG2 (Hepatoblastoma Cells) (cluster #5 Of 8), Other |
Other |
190 |
0.52 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.48 |
0.54 |
-21.38 |
5 |
8 |
0 |
130 |
253.262 |
5 |
↓
|
|
|
|
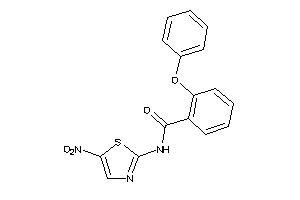
Analogs
-
5329781
-
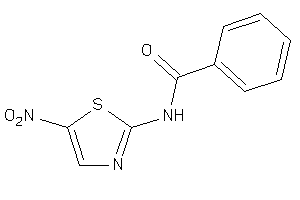
-
4578540
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
4.5 |
-15.06 |
2 |
7 |
0 |
108 |
265.25 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.45 |
5.5 |
-53.82 |
1 |
7 |
-1 |
111 |
264.242 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
1700 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
13.15 |
-14.07 |
0 |
4 |
0 |
43 |
354.816 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
14.25 |
-43.92 |
2 |
4 |
1 |
47 |
355.824 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.67 |
13.48 |
-29.56 |
1 |
4 |
1 |
45 |
355.824 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
1700 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
13.92 |
-13.8 |
0 |
4 |
0 |
43 |
354.816 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
14.26 |
-44.02 |
2 |
4 |
1 |
47 |
355.824 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
14.25 |
-30.01 |
1 |
4 |
1 |
45 |
355.824 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
6380 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
2.7 |
-10.26 |
1 |
4 |
0 |
59 |
312.365 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-2-O |
Hepatitis B Virus (cluster #2 Of 3), Other |
Other |
3500 |
0.45 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
4.81 |
-11.04 |
2 |
4 |
0 |
62 |
333.594 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.96 |
5.81 |
-49.43 |
1 |
4 |
-1 |
65 |
332.586 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.48 |
2.62 |
-41.54 |
1 |
4 |
-1 |
69 |
332.586 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-2-O |
Hepatitis B Virus (cluster #2 Of 3), Other |
Other |
760 |
0.43 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
1.5 |
-14.06 |
1 |
4 |
0 |
55 |
334.169 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-2-O |
Hepatitis B Virus (cluster #2 Of 3), Other |
Other |
660 |
0.41 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
0.08 |
-14.69 |
2 |
6 |
0 |
95 |
306.705 |
3 |
↓
|
|
|
|
Analogs
-
39914982
-
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-2-O |
Hepatitis B Virus (cluster #2 Of 3), Other |
Other |
730 |
0.43 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
1.43 |
-13.87 |
1 |
4 |
0 |
55 |
381.169 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
9100 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
12.65 |
-8.69 |
1 |
4 |
0 |
43 |
363.223 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.53 |
12.76 |
-41.1 |
2 |
4 |
1 |
47 |
364.231 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
9100 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
11.29 |
-11.47 |
1 |
4 |
0 |
43 |
363.223 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.53 |
11.43 |
-42.3 |
2 |
4 |
1 |
47 |
364.231 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
9100 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
11.31 |
-11.5 |
1 |
4 |
0 |
43 |
363.223 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.53 |
11.43 |
-42.24 |
2 |
4 |
1 |
47 |
364.231 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
9100 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
12.65 |
-8.67 |
1 |
4 |
0 |
43 |
363.223 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.53 |
13.16 |
-30.27 |
2 |
4 |
1 |
48 |
364.231 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.53 |
12.75 |
-41.06 |
2 |
4 |
1 |
47 |
364.231 |
2 |
↓
|
|
|
|
Analogs
-
631182
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
4700 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
13.05 |
-8.18 |
1 |
4 |
0 |
43 |
379.678 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.05 |
13.55 |
-29.83 |
2 |
4 |
1 |
48 |
380.686 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.05 |
13.14 |
-40.7 |
2 |
4 |
1 |
47 |
380.686 |
2 |
↓
|
|
|
|
Analogs
-
631182
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
4700 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
11.7 |
-10.71 |
1 |
4 |
0 |
43 |
379.678 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.05 |
12.48 |
-30.15 |
2 |
4 |
1 |
48 |
380.686 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.05 |
11.11 |
-41.38 |
2 |
4 |
1 |
47 |
380.686 |
2 |
↓
|
|
|
|
Analogs
-
631182
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
4700 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
11.68 |
-10.71 |
1 |
4 |
0 |
43 |
379.678 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.05 |
11.53 |
-29.65 |
2 |
4 |
1 |
48 |
380.686 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.05 |
11.13 |
-41.38 |
2 |
4 |
1 |
47 |
380.686 |
2 |
↓
|
|
|
|
Analogs
-
631182
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
4700 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.87 |
13.05 |
-8.17 |
1 |
4 |
0 |
43 |
379.678 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.05 |
13.56 |
-29.83 |
2 |
4 |
1 |
48 |
380.686 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.05 |
13.15 |
-40.71 |
2 |
4 |
1 |
47 |
380.686 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8800 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
0.63 |
-8.22 |
1 |
4 |
0 |
42 |
352.869 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8800 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
0.24 |
-10.31 |
1 |
4 |
0 |
42 |
352.869 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8800 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
0.23 |
-10.36 |
1 |
4 |
0 |
42 |
352.869 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
8800 |
0.28 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.12 |
0.64 |
-8.28 |
1 |
4 |
0 |
42 |
352.869 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
5900 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
12.2 |
-9.25 |
1 |
4 |
0 |
43 |
346.768 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.02 |
12.71 |
-31.13 |
2 |
4 |
1 |
48 |
347.776 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.02 |
12.31 |
-41.87 |
2 |
4 |
1 |
47 |
347.776 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
5900 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
10.84 |
-12.38 |
1 |
4 |
0 |
43 |
346.768 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.02 |
11.4 |
-30.55 |
2 |
4 |
1 |
48 |
347.776 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.02 |
10.98 |
-42.97 |
2 |
4 |
1 |
47 |
347.776 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
5900 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
10.86 |
-12.35 |
1 |
4 |
0 |
43 |
346.768 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.02 |
11.64 |
-31.54 |
2 |
4 |
1 |
48 |
347.776 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.02 |
10.98 |
-43.03 |
2 |
4 |
1 |
47 |
347.776 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
5900 |
0.31 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
12.2 |
-9.24 |
1 |
4 |
0 |
43 |
346.768 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.02 |
12.7 |
-31.1 |
2 |
4 |
1 |
48 |
347.776 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.02 |
12.3 |
-41.88 |
2 |
4 |
1 |
47 |
347.776 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2100 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
12.75 |
-8.59 |
1 |
4 |
0 |
43 |
407.674 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
13.27 |
-30.42 |
2 |
4 |
1 |
48 |
408.682 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.67 |
12.86 |
-41.11 |
2 |
4 |
1 |
47 |
408.682 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2100 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
11.4 |
-11.43 |
1 |
4 |
0 |
43 |
407.674 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
11.96 |
-29.93 |
2 |
4 |
1 |
48 |
408.682 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.67 |
11.53 |
-42.28 |
2 |
4 |
1 |
47 |
408.682 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2100 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
11.41 |
-11.44 |
1 |
4 |
0 |
43 |
407.674 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
12.19 |
-30.71 |
2 |
4 |
1 |
48 |
408.682 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.67 |
11.53 |
-42.3 |
2 |
4 |
1 |
47 |
408.682 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2100 |
0.33 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
12.76 |
-8.59 |
1 |
4 |
0 |
43 |
407.674 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
13.26 |
-30.41 |
2 |
4 |
1 |
48 |
408.682 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.67 |
12.85 |
-41.07 |
2 |
4 |
1 |
47 |
408.682 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
7400 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
1.25 |
-8.16 |
1 |
4 |
0 |
42 |
356.832 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
7400 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.86 |
-10.96 |
1 |
4 |
0 |
42 |
356.832 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
7400 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.86 |
-10.91 |
1 |
4 |
0 |
42 |
356.832 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
7400 |
0.29 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
1.07 |
-8.89 |
1 |
4 |
0 |
42 |
356.832 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2200 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
13.5 |
-9.06 |
1 |
4 |
0 |
43 |
356.832 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.68 |
14.58 |
-105.66 |
3 |
4 |
2 |
49 |
358.848 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
14.02 |
-30.1 |
2 |
4 |
1 |
44 |
357.84 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2200 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
12.14 |
-11.43 |
1 |
4 |
0 |
43 |
356.832 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.68 |
13.52 |
-105.27 |
3 |
4 |
2 |
49 |
358.848 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
12.66 |
-31.13 |
2 |
4 |
1 |
44 |
357.84 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
2200 |
0.32 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.50 |
12.13 |
-11.4 |
1 |
4 |
0 |
43 |
356.832 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.68 |
13.14 |
-105.35 |
3 |
4 |
2 |
49 |
358.848 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.50 |
12.65 |
-31.12 |
2 |
4 |
1 |
44 |
357.84 |
2 |
↓
|
|