|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101697-1-O |
Camelpox Virus (cluster #1 Of 1), Other |
Other |
8800 |
0.39 |
Functional ≤ 10μM
|
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
1300 |
0.46 |
Functional ≤ 10μM |
|
Z101848-1-O |
Human Herpesvirus 8 (cluster #1 Of 1), Other |
Other |
64 |
0.56 |
Functional ≤ 10μM
|
|
Z102080-1-O |
Simplexvirus (cluster #1 Of 1), Other |
Other |
3300 |
0.43 |
Functional ≤ 10μM
|
|
Z102131-1-O |
Vaccinia Virus Copenhagen (cluster #1 Of 1), Other |
Other |
9800 |
0.39 |
Functional ≤ 10μM
|
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
9500 |
0.39 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
95 |
0.55 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
480 |
0.49 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
70 |
0.56 |
Functional ≤ 10μM |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
940 |
0.47 |
Functional ≤ 10μM |
|
Z50599-1-O |
Cowpox Virus (cluster #1 Of 1), Other |
Other |
9000 |
0.39 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
9400 |
0.39 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
570 |
0.49 |
Functional ≤ 10μM
|
|
Z50604-1-O |
Murid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
40 |
0.58 |
Functional ≤ 10μM
|
|
Z50624-1-O |
Human Adenovirus Type 2 (cluster #1 Of 1), Other |
Other |
3100 |
0.43 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
10000 |
0.39 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
500 |
0.49 |
Functional ≤ 10μM
|
|
Z80709-1-O |
Osteoclast-like (Bone Marrow Cells) (cluster #1 Of 1), Other |
Other |
10000 |
0.39 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
600 |
0.48 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 1), Other |
Other |
1900 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.70 |
-5.05 |
-52.83 |
4 |
9 |
-1 |
151 |
278.181 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.35 |
-6.46 |
-25.45 |
5 |
7 |
0 |
122 |
259.287 |
2 |
↓
|
|
|
|
Analogs
-
17002582
-
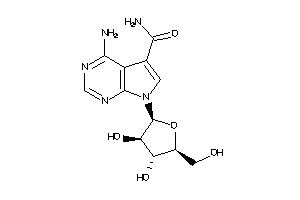
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.18 |
-13.01 |
-18.69 |
7 |
10 |
0 |
169 |
309.282 |
3 |
↓
|
|
|
|
Analogs
-
3881944
-
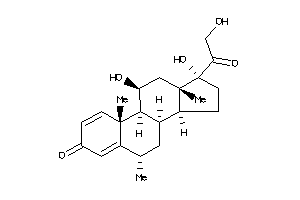
-
3881945
-
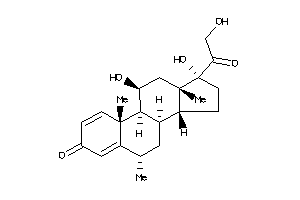
-
3881946
-
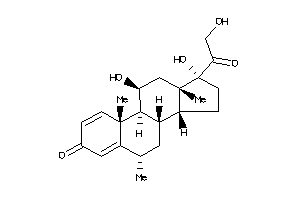
-
5275964
-
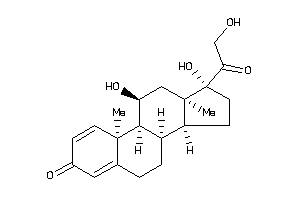
-
5276791
-
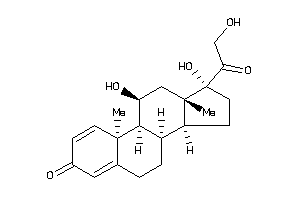
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2762 |
0.30 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2660 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.44 |
Binding ≤ 10μM
|
|
GLNA-1-E |
Glutamine Synthetase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.40 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
37 |
0.40 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Functional ≤ 10μM
|
|
MCR-2-E |
Mineralocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM |
|
Z100081-2-O |
PBMC (Peripheral Blood Mononuclear Cells) (cluster #2 Of 4), Other |
Other |
25 |
0.41 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z80110-2-O |
CV-1 (Kidney Cells) (cluster #2 Of 2), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
760 |
0.33 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z80954-3-O |
HFF (Foreskin Fibroblasts) (cluster #3 Of 4), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z81011-3-O |
Human Cell Lines (cluster #3 Of 3), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
16 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
3.09 |
-16.14 |
3 |
5 |
0 |
95 |
360.45 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.96 |
11.84 |
-46.11 |
0 |
3 |
-1 |
57 |
365.836 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
5.55 |
9.3 |
-8.83 |
2 |
3 |
0 |
58 |
366.844 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.55 |
10.22 |
-45.56 |
1 |
3 |
-1 |
60 |
365.836 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
8100 |
0.40 |
Functional ≤ 10μM |
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
60 |
0.56 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
990 |
0.47 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
5000 |
0.41 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
590 |
0.48 |
Functional ≤ 10μM
|
|
Z50529-2-O |
Cytomegalovirus (cluster #2 Of 2), Other |
Other |
8800 |
0.39 |
Functional ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
990 |
0.47 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
500 |
0.49 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
9000 |
0.39 |
Functional ≤ 10μM
|
|
Z50604-1-O |
Murid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
5500 |
0.41 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
4000 |
0.42 |
Functional ≤ 10μM
|
|
Z80253-1-O |
MEF (Embryonic Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
5000 |
0.41 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
9400 |
0.39 |
Functional ≤ 10μM
|
|
Z80823-1-O |
E6SM (Embryonic Skin-muscle Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
1 |
0.70 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
900 |
0.47 |
Functional ≤ 10μM
|
|
Z80943-1-O |
HEL 299 (cluster #1 Of 1), Other |
Other |
630 |
0.48 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8400 |
0.39 |
Functional ≤ 10μM |
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
3500 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.16 |
-1.32 |
-22.09 |
5 |
9 |
0 |
139 |
255.234 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 84 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM |
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
9600 |
0.44 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
7600 |
0.45 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
6900 |
0.45 |
Functional ≤ 10μM
|
|
Z50527-1-O |
Human Herpesvirus 3 (cluster #1 Of 3), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
4900 |
0.46 |
Functional ≤ 10μM |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
7500 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
90 |
0.62 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
384 |
0.56 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
990 |
0.53 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
20 |
0.67 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
2900 |
0.48 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
7540 |
0.45 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8100 |
0.45 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
8710 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-0.6 |
-19.69 |
4 |
8 |
0 |
119 |
225.208 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
-2.66 |
-29.41 |
2 |
5 |
0 |
71 |
390.892 |
6 |
↓
|
|
|
|
Analogs
-
13977324
-
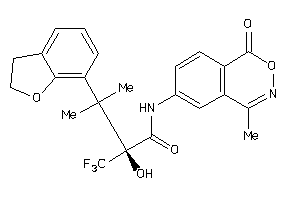
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
8.42 |
-19.84 |
2 |
7 |
0 |
102 |
476.451 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.69 |
6.44 |
-36.26 |
3 |
7 |
1 |
107 |
477.459 |
5 |
↓
|
|
|
|
Analogs
-
6238318
-
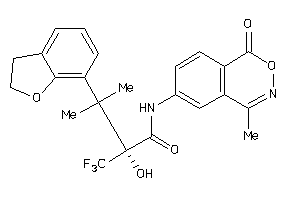
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
8.38 |
-20.96 |
2 |
7 |
0 |
102 |
476.451 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.69 |
6.34 |
-36.26 |
3 |
7 |
1 |
107 |
477.459 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
8800 |
0.39 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8800 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.80 |
-8.25 |
-15.91 |
5 |
7 |
0 |
112 |
267.314 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.40 |
-1.05 |
-232.31 |
2 |
9 |
-3 |
161 |
319.126 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.40 |
-0.33 |
-404.84 |
1 |
9 |
-4 |
164 |
318.118 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.29 |
2.71 |
-6.35 |
2 |
3 |
0 |
41 |
233.311 |
3 |
↓
|
|
|
|
Analogs
-
7992842
-
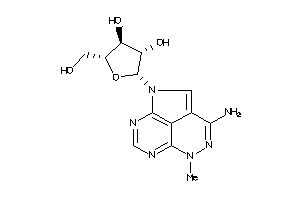
-
13099130
-
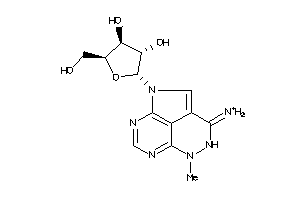
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.07 |
-3.05 |
-19.05 |
5 |
10 |
0 |
144 |
320.309 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80954-4-O |
HFF (Foreskin Fibroblasts) (cluster #4 Of 4), Other |
Other |
3300 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
2.34 |
-32.95 |
4 |
5 |
1 |
77 |
235.292 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.19 |
4.16 |
-14.57 |
3 |
5 |
0 |
72 |
234.284 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.19 |
4.01 |
-12.77 |
3 |
5 |
0 |
72 |
234.284 |
2 |
↓
|
|
|
|
Analogs
-
3978012
-
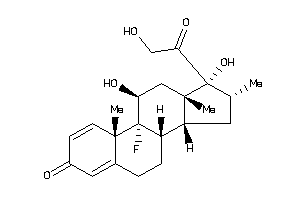
-
3978013
-
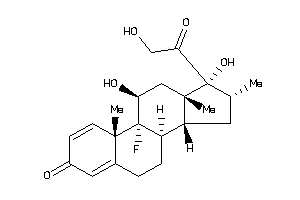
-
3978014
-
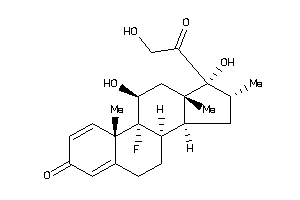
-
3978043
-
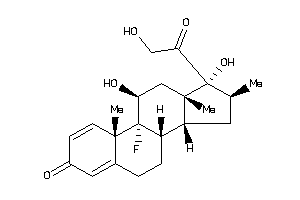
-
3978044
-
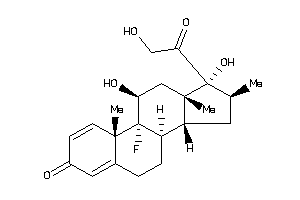
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-2-E |
Androgen Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.38 |
Binding ≤ 10μM
|
|
MCR-1-E |
Mineralocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
PRGR-1-E |
Progesterone Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Binding ≤ 10μM
|
|
GCR-2-E |
Glucocorticoid Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
20 |
0.38 |
Functional ≤ 10μM
|
|
Z80178-2-O |
J774.A1 (Macrophage Cells) (cluster #2 Of 3), Other |
Other |
48 |
0.37 |
Functional ≤ 10μM
|
|
Z80301-2-O |
N9 (cluster #2 Of 2), Other |
Other |
74 |
0.36 |
Functional ≤ 10μM
|
|
Z80418-3-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #3 Of 9), Other |
Other |
860 |
0.30 |
Functional ≤ 10μM
|
|
Z80548-4-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #4 Of 5), Other |
Other |
7 |
0.41 |
Functional ≤ 10μM
|
|
Z80954-3-O |
HFF (Foreskin Fibroblasts) (cluster #3 Of 4), Other |
Other |
2 |
0.43 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
17 |
0.39 |
Functional ≤ 10μM
|
|
Z81338-2-O |
T-cells (cluster #2 Of 3), Other |
Other |
5 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
3.98 |
-16.38 |
3 |
5 |
0 |
95 |
392.467 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
8.29 |
-30.4 |
1 |
6 |
0 |
64 |
431.945 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
6343 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.79 |
-2.11 |
-12.26 |
2 |
6 |
0 |
76 |
376.21 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.28 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.28 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8500 |
0.18 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.19 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9300 |
0.18 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.19 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.25 |
Binding ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
970 |
0.21 |
Functional ≤ 10μM
|
|
Z80046-1-O |
BT-474 (Breast Ductal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
6600 |
0.18 |
Functional ≤ 10μM
|
|
Z80300-1-O |
N87 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
4350 |
0.19 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.30 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
8500 |
0.18 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
9900 |
0.18 |
Functional ≤ 10μM
|
|
Z80961-1-O |
HN5 (Squamous Cell Carcinoma) (cluster #1 Of 1), Other |
Other |
25 |
0.27 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
6270 |
0.18 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
5900 |
0.18 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
5400 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.16 |
-3.53 |
-72.53 |
3 |
8 |
1 |
110 |
582.077 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.29 |
-3.41 |
-224 |
2 |
8 |
-3 |
157 |
280.089 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.29 |
-2.3 |
-380.72 |
1 |
8 |
-4 |
159 |
279.081 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.29 |
-4.83 |
-114.76 |
3 |
8 |
-2 |
154 |
281.097 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
3.02 |
-11.13 |
3 |
5 |
0 |
93 |
294.31 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADK-1-E |
Adenosine Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
Z50076-1-O |
Poliovirus (cluster #1 Of 2), Other |
Other |
30 |
0.55 |
Functional ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
500 |
0.46 |
Functional ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
500 |
0.46 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
60 |
0.53 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
60 |
0.53 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
40 |
0.55 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
68 |
0.53 |
Functional ≤ 10μM
|
|
Z80945-1-O |
HEp-2 (Laryngeal Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
2 |
0.64 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
400 |
0.47 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
1000 |
0.44 |
Functional ≤ 10μM
|
|
Z80193-3-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
40 |
0.55 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.93 |
-3.88 |
-15.82 |
5 |
8 |
0 |
127 |
266.257 |
2 |
↓
|
|
|
|
Analogs
-
49785288
-
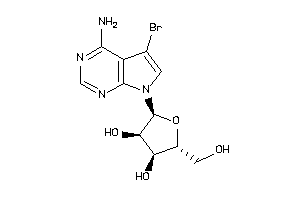
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADK-1-E |
Adenosine Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
120 |
0.48 |
Binding ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
600 |
0.44 |
Functional ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
600 |
0.44 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
4000 |
0.38 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
1200 |
0.41 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
3000 |
0.39 |
Functional ≤ 10μM
|
|
Z81032-1-O |
Human Neoplasticcell Line (cluster #1 Of 1), Other |
Other |
2300 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
-3.13 |
-12.98 |
5 |
8 |
0 |
127 |
345.153 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.14 |
5.9 |
-46.73 |
5 |
8 |
1 |
110 |
423.472 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.19 |
-7.25 |
-32.98 |
6 |
8 |
1 |
128 |
346.161 |
2 |
↓
|
|
|
|
Analogs
-
5161614
-
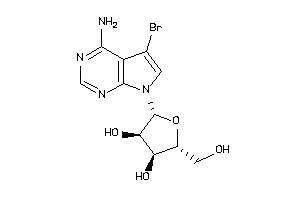
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
-2.9 |
-12.43 |
5 |
8 |
0 |
127 |
345.153 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.48 |
8.09 |
-93.83 |
5 |
6 |
2 |
87 |
425.483 |
4 |
↓
|
|
|
|
Analogs
-
5385245
-
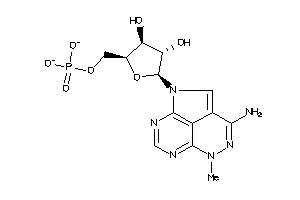
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.74 |
-8.93 |
-140.05 |
4 |
13 |
-2 |
196 |
398.272 |
4 |
↓
|
|
|
|
Analogs
-
3653422
-
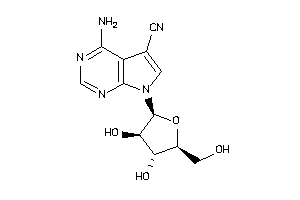
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADK-1-E |
Adenosine Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
310 |
0.43 |
Binding ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
50 |
0.49 |
Functional ≤ 10μM
|
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
50 |
0.49 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
400 |
0.43 |
Functional ≤ 10μM
|
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
5 |
0.55 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
4 |
0.56 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
10 |
0.53 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8000 |
0.34 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
30 |
0.50 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
30 |
0.50 |
Functional ≤ 10μM
|
|
Z80193-3-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #3 Of 4), Other |
Other |
4 |
0.56 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.25 |
-9.07 |
-13.99 |
5 |
9 |
0 |
150 |
291.267 |
2 |
↓
|
|
|
|
Analogs
-
5835275
-
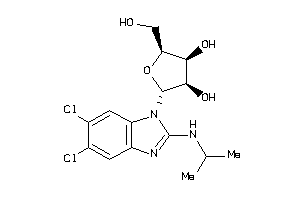
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
-9.16 |
-13.04 |
4 |
7 |
0 |
99 |
376.24 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.29 |
-8.92 |
-31.25 |
5 |
7 |
1 |
101 |
377.248 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FPPS-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
82 |
0.58 |
Binding ≤ 10μM
|
|
Q0GKD7-1-E |
Farnesyl Pyrophosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
|
Q197X6-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.59 |
Binding ≤ 10μM
|
|
Q8WS26-1-E |
Farnesyl Diphosphate Synthase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.62 |
Binding ≤ 10μM
|
|
Z50418-6-O |
Trypanosoma Brucei (cluster #6 Of 6), Other |
Other |
8600 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-17-O |
Plasmodium Falciparum (cluster #17 Of 22), Other |
Other |
1400 |
0.48 |
Functional ≤ 10μM
|
|
Z50459-5-O |
Leishmania Donovani (cluster #5 Of 8), Other |
Other |
2300 |
0.46 |
Functional ≤ 10μM
|
|
Z50472-3-O |
Toxoplasma Gondii (cluster #3 Of 4), Other |
Other |
490 |
0.52 |
Functional ≤ 10μM
|
|
Z50594-2-O |
Mus Musculus (cluster #2 Of 9), Other |
Other |
300 |
0.54 |
Functional ≤ 10μM
|
|
Z50725-4-O |
Trypanosoma Brucei Rhodesiense (cluster #4 Of 7), Other |
Other |
8600 |
0.42 |
Functional ≤ 10μM
|
|
Z50759-2-O |
Dictyostelium Discoideum (cluster #2 Of 2), Other |
Other |
3000 |
0.45 |
Functional ≤ 10μM
|
|
Z80954-2-O |
HFF (Foreskin Fibroblasts) (cluster #2 Of 4), Other |
Other |
2400 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.23 |
-3.55 |
-231.08 |
2 |
8 |
-3 |
157 |
280.089 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.23 |
-2.31 |
-381.96 |
1 |
8 |
-4 |
159 |
279.081 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.23 |
-4.53 |
-107.38 |
3 |
8 |
-2 |
154 |
281.097 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 49 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.12 |
-2.4 |
-165.36 |
3 |
9 |
-2 |
163 |
270.074 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.12 |
-1.74 |
-366.18 |
1 |
9 |
-4 |
164 |
268.058 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.12 |
-3.45 |
-71.58 |
4 |
9 |
-1 |
160 |
271.082 |
4 |
↓
|
|