|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
4800 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.60 |
8.96 |
-7.35 |
1 |
5 |
0 |
50 |
284.363 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.60 |
9.31 |
-31.21 |
2 |
5 |
1 |
52 |
285.371 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
500 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
8.18 |
-7.1 |
1 |
5 |
0 |
50 |
270.336 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.20 |
8.53 |
-31.78 |
2 |
5 |
1 |
52 |
271.344 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 99 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
8.91 |
-32.71 |
0 |
5 |
1 |
41 |
336.367 |
2 |
↓
|
|
|
|
Analogs
-
338122
-
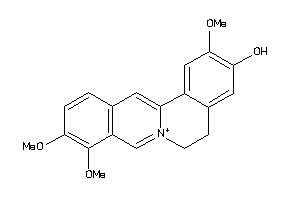
Draw
Identity
99%
90%
80%
70%
Vendors
And 42 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
400 |
0.34 |
Binding ≤ 10μM |
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.33 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
9.24 |
-33.78 |
0 |
5 |
1 |
41 |
352.41 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1800 |
0.31 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
9.11 |
-38.62 |
0 |
5 |
1 |
41 |
352.41 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
4863210
-
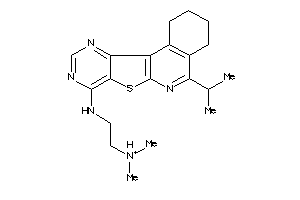
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1000 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.84 |
-2.18 |
-42.93 |
2 |
5 |
1 |
55 |
356.519 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
3279 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
1.69 |
-50.66 |
2 |
7 |
1 |
78 |
461.604 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
122 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
10.77 |
-31.07 |
4 |
5 |
1 |
78 |
364.469 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
122 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.26 |
10.74 |
-31.28 |
4 |
5 |
1 |
78 |
364.469 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
8 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
8.87 |
-25.4 |
2 |
9 |
0 |
125 |
526.582 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
4 |
0.35 |
Binding ≤ 10μM
|
|
Q9N1N9-1-E |
Butyrylcholinesterase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.27 |
9.64 |
-42.73 |
4 |
6 |
1 |
85 |
460.598 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
90 |
0.34 |
Binding ≤ 10μM
|
|
AGTRB-1-E |
Angiotensin II Type 1b (AT-1b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.76 |
13.53 |
-14.72 |
0 |
4 |
0 |
56 |
385.511 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
94 |
0.55 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
4.33 |
-56.88 |
4 |
3 |
1 |
61 |
243.33 |
0 |
↓
|
|
|
|
Analogs
-
39916778
-
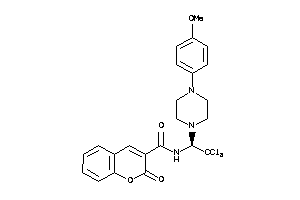
-
39916779
-
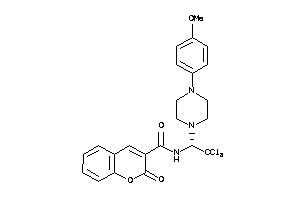
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
7500 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
-0.06 |
-24.13 |
0 |
6 |
0 |
62 |
364.401 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
540 |
0.32 |
Binding ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
153 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
4180 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
4.37 |
-47.92 |
1 |
7 |
1 |
83 |
370.381 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
9300 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
-0.04 |
-23.29 |
0 |
5 |
0 |
53 |
334.375 |
2 |
↓
|
|
|
|
Analogs
-
644561
-
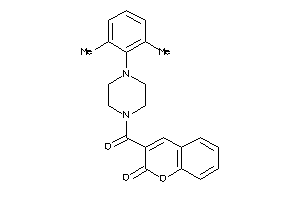
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
6700 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
0.77 |
-21.2 |
0 |
5 |
0 |
53 |
348.402 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Binding ≤ 10μM
|
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
12 |
0.44 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1560 |
0.33 |
Binding ≤ 10μM |
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2500 |
0.31 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
104 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
80 |
0.50 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
280 |
0.46 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
128 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
-0.07 |
-7.08 |
1 |
5 |
0 |
48 |
275.352 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.90 |
2.61 |
-39.01 |
2 |
5 |
1 |
50 |
276.36 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.90 |
4.54 |
-76.78 |
3 |
5 |
2 |
51 |
277.368 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
8000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.62 |
12.75 |
-11.19 |
1 |
4 |
0 |
34 |
448.441 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.29 |
12.98 |
-11.5 |
1 |
4 |
0 |
36 |
448.441 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2900 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
-0.5 |
-10.42 |
2 |
6 |
0 |
80 |
491.535 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2900 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.81 |
-0.5 |
-10.72 |
2 |
6 |
0 |
80 |
491.535 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
8.04 |
-9.39 |
2 |
3 |
0 |
53 |
347.439 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.10 |
8.76 |
-51.15 |
1 |
3 |
-1 |
56 |
346.431 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.10 |
7.6 |
-8.56 |
3 |
3 |
0 |
54 |
348.447 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
4100 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
7.99 |
-8.94 |
2 |
3 |
0 |
53 |
347.439 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.10 |
8.76 |
-51.1 |
1 |
3 |
-1 |
56 |
346.431 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.10 |
7.6 |
-8.18 |
3 |
3 |
0 |
54 |
348.447 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2500 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
5.65 |
-12.43 |
3 |
5 |
0 |
84 |
368.458 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
794 |
0.45 |
Binding ≤ 10μM
|
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
19 |
0.57 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.97 |
-1.77 |
-50.76 |
3 |
3 |
1 |
49 |
257.357 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
-0.48 |
-10.81 |
1 |
7 |
0 |
89 |
533.479 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
6700 |
0.27 |
Binding ≤ 10μM
|
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
6700 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
7.32 |
-14.4 |
2 |
6 |
0 |
88 |
404.253 |
4 |
↓
|
|
|
|
Analogs
-
22058728
-
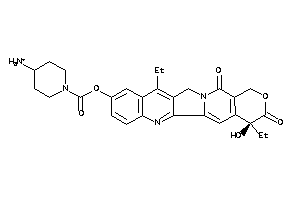
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
51 |
0.24 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
4 |
0.27 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
200 |
0.22 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
22 |
0.25 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
220 |
0.22 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
1200 |
0.19 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
9015 |
0.16 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
4 |
0.27 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
100 |
0.23 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
6528 |
0.17 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
570 |
0.20 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
9 |
0.26 |
Functional ≤ 10μM
|
|
Z81186-2-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #2 Of 2), Other |
Other |
1160 |
0.19 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
1140 |
0.19 |
Functional ≤ 10μM
|
|
Z81248-1-O |
Malme-3M (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
200 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
-1.53 |
-52.64 |
2 |
10 |
1 |
115 |
587.697 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.10 |
-1.49 |
-93.29 |
3 |
10 |
2 |
116 |
588.705 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
760 |
0.31 |
Binding ≤ 10μM
|
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
760 |
0.31 |
Binding ≤ 10μM |
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
51 |
0.36 |
Binding ≤ 10μM |
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
240 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.15 |
9.65 |
-8.23 |
1 |
5 |
0 |
45 |
379.504 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.02 |
-1.72 |
-52.77 |
1 |
5 |
1 |
58 |
491.677 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
63 |
0.56 |
Binding ≤ 10μM
|
|
CNR1-5-E |
Cannabinoid CB1 Receptor (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.66 |
Binding ≤ 10μM
|
|
FAAH1-5-E |
Anandamide Amidohydrolase (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
460 |
0.49 |
Binding ≤ 10μM
|
|
MGLL-3-E |
Monoglyceride Lipase (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
10 |
0.62 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.37 |
5.43 |
-8.31 |
0 |
5 |
0 |
58 |
334.523 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
3390 |
0.35 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
1120 |
0.38 |
Binding ≤ 10μM
|
|
AOFB-4-E |
Monoamine Oxidase B (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
2.82 |
-14.8 |
0 |
3 |
0 |
39 |
314.768 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
9887 |
0.35 |
Binding ≤ 10μM
|
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
9890 |
0.35 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2490 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
335 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
-2.69 |
-9.02 |
2 |
3 |
0 |
48 |
218.231 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
367100
-
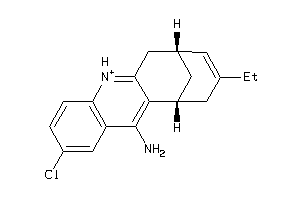
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
257 |
0.44 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
762 |
0.41 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
2150 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.77 |
9.05 |
-26.42 |
3 |
2 |
1 |
40 |
299.825 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.77 |
8.68 |
-6.24 |
2 |
2 |
0 |
39 |
298.817 |
1 |
↓
|
|
|
|
Analogs
-
367097
-
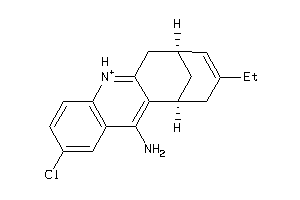
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
257 |
0.44 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
762 |
0.41 |
Binding ≤ 10μM
|
|
Z50418-2-O |
Trypanosoma Brucei (cluster #2 Of 6), Other |
Other |
2150 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.77 |
9.06 |
-26.56 |
3 |
2 |
1 |
40 |
299.825 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.77 |
8.69 |
-6.12 |
2 |
2 |
0 |
39 |
298.817 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
650 |
0.41 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
501 |
0.42 |
Binding ≤ 10μM
|
|
ATP4A-2-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.49 |
Functional ≤ 10μM
|
|
ATP4B-2-E |
Potassium-transporting ATPase Beta Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.49 |
Functional ≤ 10μM
|
|
HRH2-2-E |
Histamine H2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
33 |
0.31 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.60 |
12.3 |
-41.19 |
4 |
5 |
1 |
71 |
476.044 |
9 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7200 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
0.36 |
-51.59 |
2 |
5 |
1 |
59 |
376.48 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
3720 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.95 |
6.76 |
-25.86 |
1 |
2 |
1 |
17 |
173.239 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.95 |
6.35 |
-8.37 |
0 |
2 |
0 |
16 |
172.231 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.32 |
Binding ≤ 10μM
|
|
Q9N1N9-1-E |
Butyrylcholinesterase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.57 |
12.66 |
-37.04 |
4 |
6 |
1 |
80 |
499.679 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
5500 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.07 |
2.08 |
-39.7 |
0 |
5 |
1 |
39 |
356.442 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
95 |
0.28 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
79 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
11.76 |
-34.77 |
4 |
6 |
1 |
84 |
491.059 |
10 |
↓
|
|