|
|
Analogs
-
3786099
-
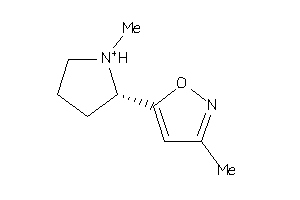
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM |
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.98 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM |
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.98 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
5.81 |
-35.61 |
1 |
3 |
1 |
30 |
167.232 |
1 |
↓
|
|
|
|
Analogs
-
8089
-
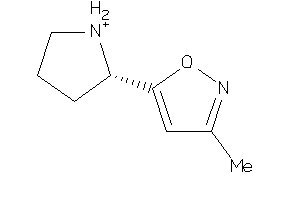
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
333 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
3.56 |
-40.1 |
2 |
3 |
1 |
43 |
153.205 |
1 |
↓
|
|
|
|
Analogs
-
13735584
-
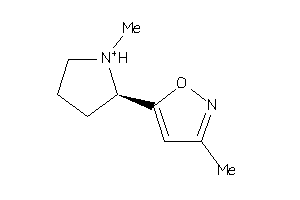
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
62 |
0.84 |
Binding ≤ 10μM |
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
62 |
0.84 |
Binding ≤ 10μM |
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
56 |
0.85 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
53 |
0.85 |
Binding ≤ 10μM |
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
4 |
0.98 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
8 |
0.94 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
380 |
0.75 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
4.98 |
-37.04 |
1 |
3 |
1 |
30 |
167.232 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
206 |
0.78 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
206 |
0.78 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
3.49 |
-29 |
1 |
3 |
1 |
30 |
162.216 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.19 |
3.77 |
-89.29 |
2 |
3 |
2 |
31 |
163.224 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
4.36 |
-40.25 |
2 |
3 |
1 |
43 |
167.232 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Binding ≤ 10μM
|
|
ACM2-6-E |
Muscarinic Acetylcholine Receptor M2 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
9150 |
0.64 |
Binding ≤ 10μM |
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5700 |
0.67 |
Binding ≤ 10μM
|
|
ACM4-6-E |
Muscarinic Acetylcholine Receptor M4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.79 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM3-3-E |
Muscarinic Acetylcholine Receptor M3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
270 |
0.84 |
Binding ≤ 10μM
|
|
Z104303-5-O |
Muscarinic Acetylcholine Receptor (cluster #5 Of 7), Other |
Other |
77 |
0.91 |
Binding ≤ 10μM |
|
Z50512-4-O |
Cavia Porcellus (cluster #4 Of 7), Other |
Other |
600 |
0.79 |
Functional ≤ 10μM
|
|
Z50592-8-O |
Oryctolagus Cuniculus (cluster #8 Of 8), Other |
Other |
545 |
0.80 |
Functional ≤ 10μM |
|
Z80024-2-O |
A9 (Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1200 |
0.75 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
8090
-
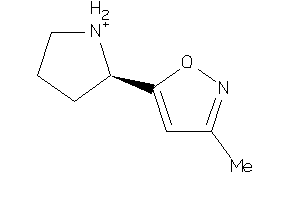
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
3.6 |
-40.56 |
2 |
3 |
1 |
43 |
153.205 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA-1-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA2-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA5-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA6-1-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHA9-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB2-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB3-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
|
ACHB4-1-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
552 |
0.73 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
4.33 |
-39.72 |
2 |
3 |
1 |
43 |
167.232 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5010 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
6.06 |
-31.2 |
1 |
3 |
1 |
30 |
202.281 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.44 |
6.34 |
-83.27 |
2 |
3 |
2 |
31 |
203.289 |
3 |
↓
|
|