|
|
|
|
|
Analogs
-
39048783
-
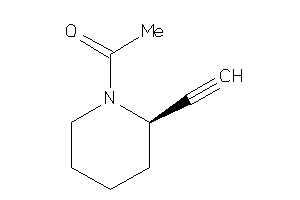
-
39048785
-
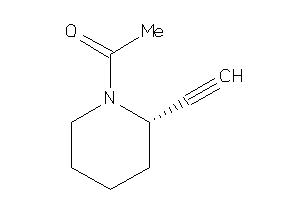
-
5160057
-
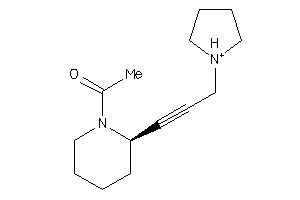
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
97 |
0.58 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.64 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
44 |
0.61 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
97 |
0.58 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
97 |
0.58 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.58 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
1.06 |
-28.79 |
1 |
3 |
1 |
24 |
235.351 |
1 |
↓
|
|
|
|
Analogs
-
6382231
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-8-E |
Acetylcholinesterase (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
8 |
1.13 |
Binding ≤ 10μM
|
|
ACHA2-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
640 |
0.87 |
Binding ≤ 10μM
|
|
ACHB2-3-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
640 |
0.87 |
Binding ≤ 10μM
|
|
ACHB4-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
640 |
0.87 |
Binding ≤ 10μM
|
|
ACHD-2-E |
Acetylcholine Receptor Protein Delta Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.89 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
739 |
0.86 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
8 |
1.13 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9100 |
0.71 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5500 |
0.74 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4300 |
0.75 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
620 |
0.87 |
Functional ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9900 |
0.70 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
230 |
0.93 |
Functional ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8900 |
0.71 |
Functional ≤ 10μM
|
|
Z104281-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #1 Of 2), Other |
Other |
1100 |
0.83 |
Binding ≤ 10μM
|
|
Z104286-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #1 Of 2), Other |
Other |
210 |
0.93 |
Binding ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 3), Other |
Other |
4800 |
0.74 |
Binding ≤ 10μM
|
|
Z104289-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 2), Other |
Other |
1300 |
0.82 |
Binding ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
590 |
0.87 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
9000 |
0.71 |
Binding ≤ 10μM
|
|
Z102306-2-O |
Aorta (cluster #2 Of 6), Other |
Other |
790 |
0.85 |
Functional ≤ 10μM
|
|
Z50512-2-O |
Cavia Porcellus (cluster #2 Of 7), Other |
Other |
500 |
0.88 |
Functional ≤ 10μM
|
|
Z50592-4-O |
Oryctolagus Cuniculus (cluster #4 Of 8), Other |
Other |
460 |
0.89 |
Functional ≤ 10μM
|
|
Z80024-2-O |
A9 (Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
100 |
0.98 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.14 |
1.5 |
-35.46 |
2 |
4 |
1 |
52 |
147.198 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
330 |
0.65 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.78 |
Functional ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.78 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.78 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.78 |
Functional ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.42 |
2.59 |
-36.97 |
0 |
3 |
1 |
20 |
193.27 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4466 |
0.62 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
8260 |
0.59 |
Binding ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
280 |
0.76 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
5.73 |
-35.78 |
1 |
3 |
1 |
31 |
170.232 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
510 |
0.73 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4466 |
0.62 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
8260 |
0.59 |
Binding ≤ 10μM
|
|
Z50597-11-O |
Rattus Norvegicus (cluster #11 Of 12), Other |
Other |
280 |
0.76 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.04 |
5.73 |
-35.74 |
1 |
3 |
1 |
31 |
170.232 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
7240 |
0.38 |
Binding ≤ 10μM |
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5304 |
0.39 |
Binding ≤ 10μM |
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2399 |
0.41 |
Binding ≤ 10μM
|
|
ACM4-4-E |
Muscarinic Acetylcholine Receptor M4 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
9120 |
0.37 |
Binding ≤ 10μM
|
|
ACM5-4-E |
Muscarinic Acetylcholine Receptor M5 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3802 |
0.40 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.40 |
Functional ≤ 10μM |
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.38 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
64 |
0.53 |
Binding ≤ 10μM |
|
Z50592-4-O |
Oryctolagus Cuniculus (cluster #4 Of 8), Other |
Other |
659 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.83 |
8.93 |
-38.89 |
1 |
4 |
1 |
38 |
281.763 |
4 |
↓
|
|
|
|
Analogs
-
2146111
-
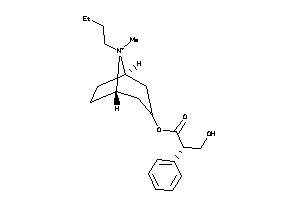
-
4214170
-
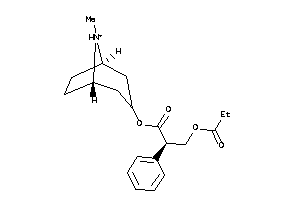
Draw
Identity
99%
90%
80%
70%
Vendors
And 29 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.47 |
9.34 |
-34.51 |
1 |
4 |
1 |
47 |
332.464 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
170 |
0.68 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
180 |
0.67 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
60 |
0.72 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
90 |
0.70 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
310 |
0.65 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
470 |
0.63 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
2.57 |
-4.94 |
0 |
4 |
0 |
49 |
193.25 |
2 |
↓
|
|
|
|
Analogs
-
3642651
-
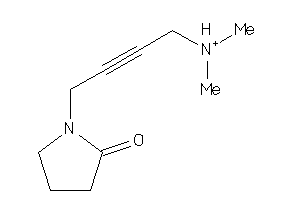
-
1530795
-
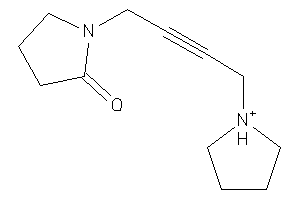
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
700 |
0.62 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
910 |
0.60 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1800 |
0.57 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2300 |
0.56 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
13 |
0.79 |
Binding ≤ 10μM
|
|
Z50512-2-O |
Cavia Porcellus (cluster #2 Of 7), Other |
Other |
4300 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.28 |
8 |
-35.62 |
0 |
3 |
1 |
20 |
195.286 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
56 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
7.78 |
-37.55 |
1 |
3 |
1 |
25 |
221.324 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
93 |
0.62 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
56 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
7.98 |
-38.11 |
1 |
3 |
1 |
25 |
221.324 |
2 |
↓
|
|
|
|
Analogs
-
1547932
-
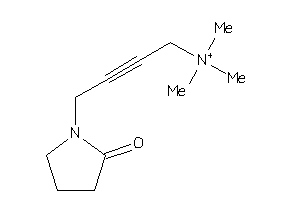
-
3642651
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-5-E |
Acetylcholinesterase (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
1500 |
0.54 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
270 |
0.61 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1500 |
0.54 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
930 |
0.56 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
5 |
0.77 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
930 |
0.56 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.66 |
Functional ≤ 10μM |
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.68 |
Functional ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
93 |
0.66 |
Binding ≤ 10μM
|
|
Z50512-2-O |
Cavia Porcellus (cluster #2 Of 7), Other |
Other |
44 |
0.69 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
7.27 |
-38.95 |
1 |
3 |
1 |
25 |
207.297 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.10 |
4.79 |
-10.78 |
0 |
3 |
0 |
24 |
206.289 |
2 |
↓
|
|