|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.04 |
2.29 |
-156.31 |
9 |
8 |
3 |
89 |
505.82 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.94 |
7.04 |
-11.84 |
1 |
10 |
0 |
126 |
459.499 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
5.85 |
-47.99 |
4 |
5 |
1 |
72 |
350.49 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.26 |
6.58 |
-83.64 |
5 |
5 |
2 |
74 |
351.498 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.26 |
7.64 |
-159.81 |
6 |
5 |
3 |
75 |
352.506 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
6.23 |
-51.25 |
4 |
5 |
1 |
72 |
350.49 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.26 |
6.94 |
-96.56 |
5 |
5 |
2 |
74 |
351.498 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.26 |
7.64 |
-163.81 |
6 |
5 |
3 |
75 |
352.506 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
193 |
0.30 |
Binding ≤ 10μM
|
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.37 |
Functional ≤ 10μM
|
|
ENV-1-V |
Envelope Glycoprotein Gp160 (cluster #1 Of 2), Viral |
Viruses |
2 |
0.39 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
9.78 |
-81.92 |
3 |
7 |
2 |
63 |
422.577 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.62 |
6.86 |
-13.96 |
1 |
7 |
0 |
60 |
420.561 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.62 |
7.56 |
-73.43 |
3 |
7 |
2 |
63 |
422.577 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
1 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
16.04 |
-233.28 |
5 |
7 |
4 |
61 |
483.749 |
17 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.93 |
13.05 |
-71.47 |
3 |
7 |
2 |
59 |
481.733 |
17 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.93 |
13.54 |
-40.73 |
2 |
7 |
1 |
57 |
480.725 |
17 |
↓
|
|
|
|
Analogs
-
37996312
-
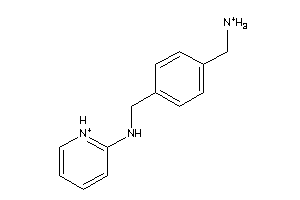
-
37998969
-
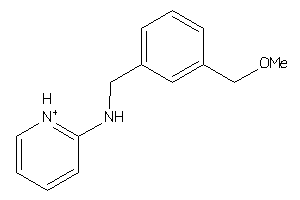
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.55 |
Functional ≤ 10μM |
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
5 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
8.37 |
-34.67 |
3 |
4 |
1 |
51 |
291.378 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.51 |
2.82 |
-60.28 |
3 |
13 |
-1 |
183 |
531.446 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.65 |
8.84 |
-71.14 |
4 |
4 |
2 |
52 |
292.386 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
112 |
0.32 |
Binding ≤ 10μM
|
|
CXCR4-1-E |
C-X-C Chemokine Receptor Type 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.37 |
Functional ≤ 10μM
|
|
Z50607-2-O |
Human Immunodeficiency Virus 1 (cluster #2 Of 10), Other |
Other |
9 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
0.24 |
5.75 |
-168.11 |
7 |
6 |
3 |
75 |
413.634 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.24 |
4.34 |
-107.26 |
6 |
6 |
2 |
70 |
412.626 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.24 |
5.03 |
-177.16 |
7 |
6 |
3 |
78 |
413.634 |
6 |
↓
|
|