|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.34 |
Binding ≤ 10μM |
|
DRD4-3-E |
Dopamine D4 Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
97 |
0.45 |
Binding ≤ 10μM |
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
240 |
0.42 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.76 |
9 |
-46.33 |
1 |
3 |
1 |
30 |
291.374 |
3 |
↓
|
|
|
|
Analogs
-
13742643
-
Draw
Identity
99%
90%
80%
70%
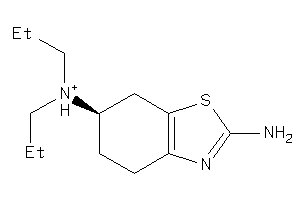
Popular Name:
2,6-Benzothiazolediamine, 4,5,6,7-tetrahydro-N6,N6-dipropyl-
2,6-Benzothiazolediamine, 4,5,6,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
57 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
7.1 |
-38.59 |
3 |
3 |
1 |
43 |
254.423 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.22 |
7.34 |
-84.04 |
4 |
3 |
2 |
45 |
255.431 |
5 |
↓
|
|
|
|
Analogs
-
13742644
-
Draw
Identity
99%
90%
80%
70%
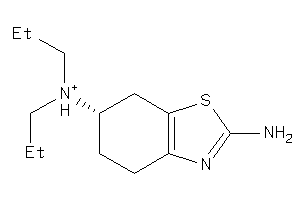
Popular Name:
2,6-Benzothiazolediamine, 4,5,6,7-tetrahydro-N6,N6-dipropyl-
2,6-Benzothiazolediamine, 4,5,6,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.72 |
Binding ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
57 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
7.02 |
-38.65 |
3 |
3 |
1 |
43 |
254.423 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.22 |
7.25 |
-84.19 |
4 |
3 |
2 |
45 |
255.431 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.47 |
Binding ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
958 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
6.02 |
-33.23 |
3 |
3 |
1 |
43 |
224.353 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.99 |
6.23 |
-86.54 |
4 |
3 |
2 |
45 |
225.361 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
-3.83 |
-44.21 |
4 |
3 |
1 |
55 |
212.342 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.09 |
-3.76 |
-87.99 |
5 |
3 |
2 |
56 |
213.35 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 67 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.74 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.85 |
Binding ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Functional ≤ 10μM
|
|
DRD4-1-E |
Dopamine D4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.53 |
Functional ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
3.81 |
-44.37 |
4 |
3 |
1 |
56 |
212.342 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.09 |
4.24 |
-88 |
5 |
3 |
2 |
57 |
213.35 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-4-E |
Alpha-2a Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.76 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.76 |
Binding ≤ 10μM
|
|
ADA2C-4-E |
Alpha-2c Adrenergic Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.76 |
Binding ≤ 10μM
|
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
6 |
0.82 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.85 |
5.05 |
-37.51 |
3 |
3 |
1 |
43 |
210.326 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.85 |
3.29 |
-5.22 |
2 |
3 |
0 |
42 |
209.318 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD2-22-E |
Dopamine D2 Receptor (cluster #22 Of 24), Eukaryotic |
Eukaryotes |
440 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
3.52 |
-5.94 |
2 |
3 |
0 |
42 |
221.329 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.75 |
3.88 |
-27.11 |
3 |
3 |
1 |
43 |
222.337 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.75 |
6.07 |
-86.72 |
4 |
3 |
2 |
45 |
223.345 |
3 |
↓
|
|