|
|
Analogs
-
44831797
-
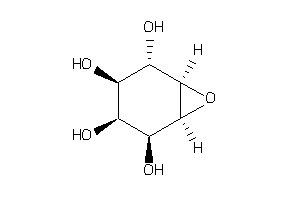
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8900 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.15 |
-9.59 |
-15.16 |
4 |
5 |
0 |
93 |
162.141 |
0 |
↓
|
|
|
|
Analogs
-
5766623
-
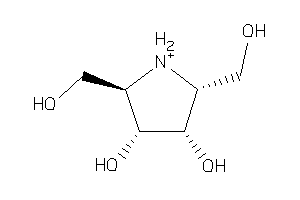
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.85 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.62 |
-11.64 |
-30.84 |
6 |
5 |
1 |
97 |
164.181 |
2 |
↓
|
|
|
|
Analogs
-
899007
-
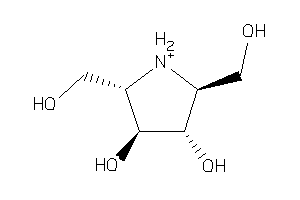
-
1492230
-
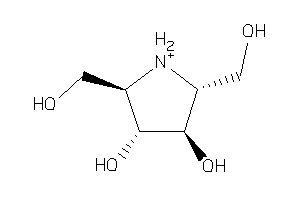
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AGAL-1-E |
Alpha-galactosidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
780 |
0.78 |
Binding ≤ 10μM
|
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.85 |
Binding ≤ 10μM
|
|
LPH-1-E |
Lactase-glycosylceramidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.62 |
-10.88 |
-32.12 |
6 |
5 |
1 |
97 |
164.181 |
2 |
↓
|
|
|
|
Analogs
-
33854099
-
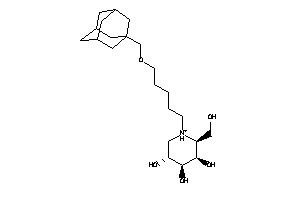
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CEGT-1-E |
Ceramide Glucosyltransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
150 |
0.34 |
Binding ≤ 10μM
|
|
GBA2-1-E |
Beta-glucosidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
30 |
0.38 |
Binding ≤ 10μM |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
2.91 |
-31.36 |
5 |
6 |
1 |
95 |
398.564 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.81 |
3.44 |
-31.92 |
4 |
6 |
0 |
97 |
397.556 |
9 |
↓
|
|
|
|
Analogs
-
6613627
-
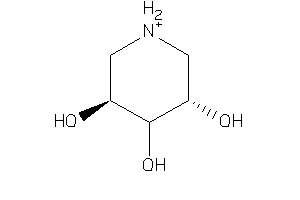
-
6613653
-
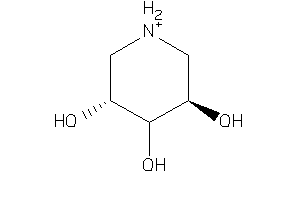
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.88 |
Binding ≤ 10μM
|
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8800 |
0.79 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-6.22 |
-45.57 |
5 |
4 |
1 |
77 |
134.155 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
3995890
-
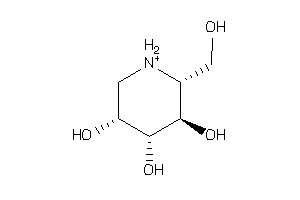
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WC34-1-E |
Alpha-glucosidase II Beta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.68 |
Binding ≤ 10μM
|
|
D3ZAN3-1-E |
Alpha-glucosidase II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.68 |
Binding ≤ 10μM
|
|
GANAB-1-E |
Neutral Alpha-glucosidase AB (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.86 |
Binding ≤ 10μM |
|
GANC-1-E |
Neutral Alpha-glucosidase C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.83 |
Binding ≤ 10μM
|
|
GDE-1-E |
Glycogen Debranching Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.86 |
Binding ≤ 10μM
|
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.73 |
Binding ≤ 10μM |
|
LYAG-1-E |
Alpha-glucosidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.81 |
Binding ≤ 10μM
|
|
MGA-1-E |
Maltase-glucoamylase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.94 |
Binding ≤ 10μM
|
|
Q43014-2-E |
Beta-glucosidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.64 |
Binding ≤ 10μM |
|
Q9LGC6-1-E |
Alpha-glucosidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.93 |
Binding ≤ 10μM
|
|
SUIS-1-E |
Sucrase-isomaltase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
650 |
0.79 |
Binding ≤ 10μM |
|
P94451-1-B |
Alpha-glucosidase (cluster #1 Of 1), Bacterial |
Bacteria |
1670 |
0.74 |
Binding ≤ 10μM
|
|
Z81057-3-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #3 Of 4), Other |
Other |
2000 |
0.73 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.40 |
-9.17 |
-39.08 |
6 |
5 |
1 |
98 |
164.181 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
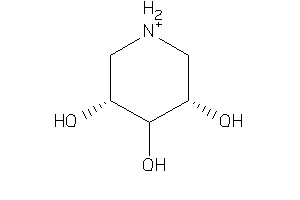
Analogs
-
6613653
-

-
14658010
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8800 |
0.79 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-8.68 |
-45.9 |
5 |
4 |
1 |
77 |
134.155 |
0 |
↓
|
|
|
|
Analogs
-
14658010
-

-
6613627
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8800 |
0.79 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
-8.68 |
-45.92 |
5 |
4 |
1 |
77 |
134.155 |
0 |
↓
|
|
|
|
|
|
|
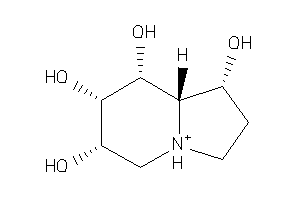
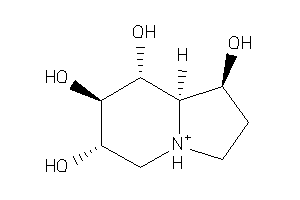
Analogs
-
1612534
-
-
3775177
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,6S,7R,8R,8aR)-1,2,3,5,6,7,8,8a-octahydroindolizine-1,6,7,8-tetrol
(1R,6S,7R,8R,8aR)-1,2,3,5,6,7,8,…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GANAB-1-E |
Neutral Alpha-glucosidase AB (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.84 |
Binding ≤ 10μM |
|
GLCM-2-E |
Glucosylceramidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.65 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.04 |
-6.74 |
-39.81 |
5 |
5 |
1 |
85 |
190.219 |
0 |
↓
|
|
|
|
|
|
|
|