|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
720 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
5.16 |
-7.87 |
0 |
3 |
0 |
43 |
188.182 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
214 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
1.87 |
-8.77 |
0 |
3 |
0 |
43 |
240.258 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
280 |
0.66 |
Binding ≤ 10μM
|
|
Q4U254-1-V |
Human Rhinovirus A Protease (cluster #1 Of 3), Viral |
Viruses |
5300 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
6.03 |
-5.42 |
0 |
2 |
0 |
34 |
227.046 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
6.52 |
-10.98 |
2 |
5 |
0 |
76 |
363.413 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.75 |
7.57 |
-42.43 |
3 |
5 |
1 |
80 |
364.421 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.74 |
-12.38 |
2 |
5 |
0 |
76 |
363.413 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.75 |
8.46 |
-44.6 |
3 |
5 |
1 |
80 |
364.421 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.23 |
-9.3 |
2 |
5 |
0 |
76 |
363.413 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.75 |
8.5 |
-45.98 |
3 |
5 |
1 |
80 |
364.421 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.64 |
-12.76 |
2 |
5 |
0 |
76 |
363.413 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.75 |
8.32 |
-47.27 |
3 |
5 |
1 |
80 |
364.421 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
3.54 |
-8.75 |
1 |
3 |
0 |
54 |
174.155 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
4.54 |
-46.63 |
0 |
3 |
-1 |
57 |
173.147 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.95 |
1.79 |
-8.21 |
0 |
3 |
0 |
43 |
242.274 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
190 |
0.86 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
0.12 |
-9.05 |
1 |
3 |
0 |
49 |
147.133 |
0 |
↓
|
|
|
|
Analogs
-
39102
-
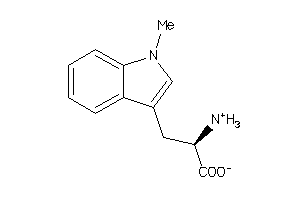
-
44197468
-
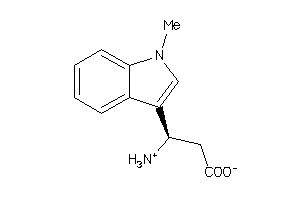
-
44197471
-
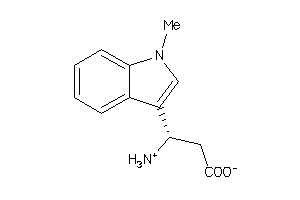
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7000 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.01 |
-1.22 |
-36.38 |
3 |
4 |
0 |
72 |
218.256 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
6.09 |
-12.34 |
0 |
2 |
0 |
34 |
158.156 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS1-2-E |
Dual Specificity Protein Phosphatase 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8450 |
0.59 |
Binding ≤ 10μM
|
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
990 |
0.70 |
Binding ≤ 10μM
|
|
MP2K1-3-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
380 |
0.75 |
Binding ≤ 10μM
|
|
MPIP2-1-E |
Dual Specificity Phosphatase Cdc25B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2760 |
0.65 |
Binding ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
127 |
0.80 |
Functional ≤ 10μM
|
|
Z81034-10-O |
A2780 (Ovarian Carcinoma Cells) (cluster #10 Of 10), Other |
Other |
2300 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
5.04 |
-6.65 |
0 |
2 |
0 |
34 |
158.156 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.92 |
-7.49 |
4 |
7 |
0 |
110 |
233.231 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.05 |
2.11 |
-32.63 |
3 |
7 |
-1 |
112 |
232.223 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.93 |
3.72 |
-37.78 |
3 |
7 |
-1 |
107 |
232.223 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
I23O1-1-E |
Indoleamine 2,3-dioxygenase 1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1100 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.33 |
1.48 |
-6.45 |
4 |
7 |
0 |
110 |
233.231 |
4 |
↓
|
|
|
|
Analogs
-
3063275
-
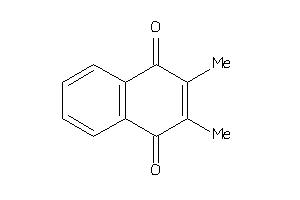
Draw
Identity
99%
90%
80%
70%
Vendors
And 56 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
5.73 |
-6.36 |
0 |
2 |
0 |
34 |
172.183 |
0 |
↓
|
|