|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
60 |
0.51 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Functional ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.44 |
Functional ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
-3.56 |
-57.45 |
3 |
5 |
-1 |
100 |
271.248 |
3 |
↓
|
|
|
|
Analogs
-
38159104
-
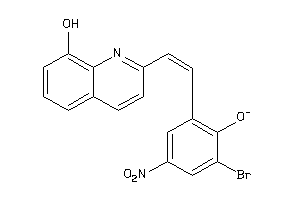
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
-2.51 |
-41.95 |
1 |
6 |
-1 |
102 |
386.181 |
3 |
↓
|
|
|
|
Analogs
-
9720706
-
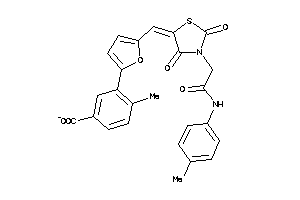
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.27 |
11.08 |
-63.62 |
1 |
8 |
-1 |
121 |
475.502 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
-1.25 |
-12.76 |
2 |
7 |
0 |
106 |
298.349 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
150 |
0.60 |
Binding ≤ 10μM
|
|
FYN-4-E |
Tyrosine-protein Kinase FYN (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
150 |
0.60 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
35 |
0.65 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9300 |
0.44 |
Binding ≤ 10μM
|
|
P85B-2-E |
PI3-kinase P85-beta Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9300 |
0.44 |
Binding ≤ 10μM
|
|
PLCG1-1-E |
Phospholipase C-gamma-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.46 |
Binding ≤ 10μM
|
|
PLCG2-2-E |
Phospholipase C-gamma-2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4900 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
3.47 |
-46.37 |
1 |
5 |
-1 |
91 |
219.172 |
2 |
↓
|
|
|
|
Analogs
-
37130512
-
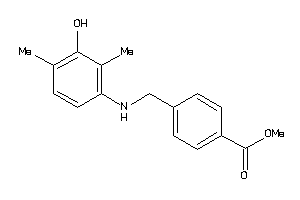
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.45 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.45 |
Binding ≤ 10μM
|
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
-4.45 |
-11.96 |
4 |
6 |
0 |
99 |
289.287 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 65 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FYN-3-E |
Tyrosine-protein Kinase FYN (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
700 |
1.08 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
260 |
1.15 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.10 |
3.04 |
-50.35 |
0 |
2 |
-1 |
40 |
127.144 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FYN-3-E |
Tyrosine-protein Kinase FYN (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
60 |
0.63 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
220 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.32 |
-6.39 |
-175.52 |
0 |
7 |
-3 |
137 |
243.172 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
620 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
6.33 |
-10.56 |
1 |
5 |
0 |
81 |
282.251 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.34 |
7.35 |
-49.07 |
0 |
5 |
-1 |
84 |
281.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
370 |
0.53 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
6.52 |
-44.95 |
0 |
5 |
-1 |
80 |
233.199 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
466 |
0.29 |
Binding ≤ 10μM
|
|
AURKA-2-E |
Serine/threonine-protein Kinase Aurora-A (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.26 |
Binding ≤ 10μM
|
|
AURKB-2-E |
Serine/threonine-protein Kinase Aurora-B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
118 |
0.31 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
396 |
0.29 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
330 |
0.29 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.34 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.41 |
Binding ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
170 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
9.24 |
-15.9 |
1 |
6 |
0 |
70 |
414.461 |
6 |
↓
|
|
|
|
Analogs
-
160657
-
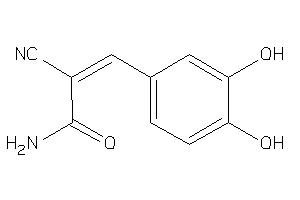
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.48 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.15 |
-2.32 |
-12.35 |
4 |
5 |
0 |
107 |
204.185 |
2 |
↓
|
|
|
|
Analogs
-
34541434
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.52 |
Binding ≤ 10μM
|
|
LUXP-1-B |
Autoinducer 2-binding Periplasmic Protein LuxP (cluster #1 Of 2), Bacterial |
Bacteria |
3000 |
0.52 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
6310 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.91 |
-0.8 |
-14.91 |
3 |
5 |
0 |
108 |
202.169 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.91 |
0.23 |
-47.75 |
2 |
5 |
-1 |
111 |
201.161 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.91 |
0.11 |
-52.62 |
2 |
5 |
-1 |
111 |
201.161 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
44199139
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FYN-4-E |
Tyrosine-protein Kinase FYN (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.63 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
240 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
0.41 |
-47.06 |
0 |
4 |
-1 |
58 |
260.063 |
3 |
↓
|
|
|
|
Analogs
-
39121418
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
1.03 |
-13.2 |
2 |
5 |
0 |
80 |
219.196 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.58 |
1.79 |
-37.02 |
1 |
5 |
-1 |
82 |
218.188 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AURKA-1-E |
Serine/threonine-protein Kinase Aurora-A (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
371 |
0.26 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
18 |
0.31 |
Binding ≤ 10μM
|
|
CSF1R-1-E |
Macrophage Colony-stimulating Factor 1 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
55 |
0.29 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
38 |
0.30 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
450 |
0.25 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
580 |
0.25 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
Z80005-2-O |
3T3 (Fibroblast Cells) (cluster #2 Of 2), Other |
Other |
25 |
0.30 |
Functional ≤ 10μM
|
|
Z80335-1-O |
NHDF (cluster #1 Of 2), Other |
Other |
25 |
0.30 |
Functional ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
8 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.25 |
11.79 |
-16.6 |
1 |
6 |
0 |
70 |
484.939 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.59 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
-2.75 |
-15.3 |
4 |
5 |
0 |
96 |
204.185 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.56 |
-1.98 |
-38.07 |
3 |
5 |
-1 |
99 |
203.177 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
763 |
0.33 |
Binding ≤ 10μM |
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
303 |
0.35 |
Binding ≤ 10μM |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
750 |
0.33 |
Binding ≤ 10μM
|
|
MAPK5-1-E |
MAP Kinase-activated Protein Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.33 |
Binding ≤ 10μM |
|
MK01-1-E |
Mitogen-activated Protein Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
950 |
0.32 |
Binding ≤ 10μM |
|
MP2K2-1-E |
Dual Specificity Mitogen-activated Protein Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.42 |
Binding ≤ 10μM |
|
TYK2-1-E |
Tyrosine-protein Kinase TYK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4100 |
0.29 |
Binding ≤ 10μM |
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
57 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
2.55 |
-11.63 |
3 |
7 |
0 |
113 |
364.394 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.53 |
3.55 |
-52.89 |
2 |
7 |
-1 |
116 |
363.386 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.53 |
3.38 |
-51.25 |
2 |
7 |
-1 |
116 |
363.386 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FGFR1-1-E |
Fibroblast Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
69 |
0.25 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.26 |
Binding ≤ 10μM
|
|
FGFR3-1-E |
Fibroblast Growth Factor Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
108 |
0.24 |
Binding ≤ 10μM
|
|
FGFR4-1-E |
Fibroblast Growth Factor Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
610 |
0.22 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.27 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.27 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
195 |
0.23 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
59 |
0.25 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.25 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
156 |
0.24 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
34 |
0.26 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.29 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.29 |
Binding ≤ 10μM
|
|
Z81057-1-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #1 Of 4), Other |
Other |
9 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
11.6 |
-47.6 |
3 |
9 |
1 |
99 |
540.644 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
4.16 |
11.94 |
-48.76 |
3 |
9 |
1 |
99 |
540.644 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
4.62 |
8.82 |
-50.42 |
3 |
9 |
1 |
99 |
540.644 |
8 |
↓
|
|
|
|
Analogs
-
5552326
-
-
14505445
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.39 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
10000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
-1.28 |
-10.92 |
4 |
4 |
0 |
81 |
244.246 |
2 |
↓
|
|
|
|
Analogs
-
6044763
-
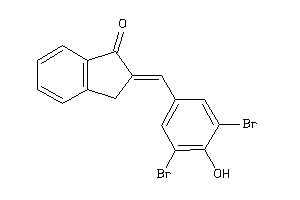
-
33686957
-
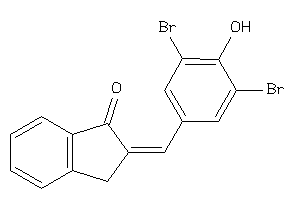
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
740 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
-1.53 |
-9.03 |
1 |
3 |
0 |
54 |
408.045 |
1 |
↓
|
|
|
|
Analogs
-
1736134
-
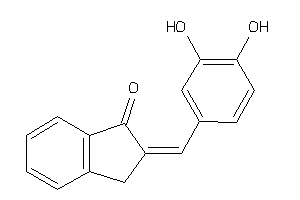
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.37 |
Binding ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
3000 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
-1.74 |
-10.87 |
2 |
4 |
0 |
74 |
266.252 |
1 |
↓
|
|
|
|
Analogs
-
36885004
-

-
37993347
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FYN-4-E |
Tyrosine-protein Kinase FYN (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
240 |
0.58 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
290 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
4.35 |
-39.27 |
1 |
7 |
-1 |
115 |
223.164 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
BRAF-1-E |
Serine/threonine-protein Kinase B-raf (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.32 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
570 |
0.30 |
Binding ≤ 10μM
|
|
MK11-1-E |
MAP Kinase P38 Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM
|
|
MK14-2-E |
MAP Kinase P38 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM
|
|
RAF1-1-E |
Serine/threonine-protein Kinase RAF (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
6.96 |
-15.54 |
3 |
6 |
0 |
82 |
389.455 |
5 |
↓
|
|