|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPB-1-E |
Aminopeptidase B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM |
|
AMPE-2-E |
Aminopeptidase A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
|
AMPN-2-E |
Aminopeptidase N (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
AMPN-2-E |
Aminopeptidase N (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
380 |
0.43 |
Binding ≤ 10μM
|
|
DPP2-2-E |
Dipeptidyl Peptidase II (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP3-1-E |
Dipeptidyl Peptidase III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP3-1-E |
Dipeptidyl Peptidase III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
DPP4-2-E |
Dipeptidyl Peptidase IV (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
|
DPP6-1-E |
Dipeptidyl Peptidase VI (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.60 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
LKHA4-1-E |
Leukotriene A4 Hydrolase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.55 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.58 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.54 |
Binding ≤ 10μM
|
|
THER-1-B |
Thermolysin (cluster #1 Of 3), Bacterial |
Bacteria |
3 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
3.01 |
-61.22 |
3 |
7 |
-1 |
119 |
293.299 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.23 |
4.26 |
-116.92 |
2 |
7 |
-2 |
121 |
292.291 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.43 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.18 |
-3.2 |
-61.08 |
1 |
6 |
-1 |
89 |
407.537 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.18 |
-4.16 |
-118.31 |
1 |
6 |
-2 |
89 |
406.529 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.27 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.58 |
0.64 |
-66.62 |
1 |
8 |
-1 |
113 |
514.639 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
-1.01 |
-107.5 |
1 |
8 |
-2 |
127 |
397.468 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.23 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
THER-1-B |
Thermolysin (cluster #1 Of 3), Bacterial |
Bacteria |
33 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.56 |
-1.57 |
-111.49 |
6 |
13 |
-2 |
220 |
541.494 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.56 |
-0.31 |
-60.51 |
7 |
13 |
-1 |
221 |
542.502 |
11 |
↓
|
|
|
|
|
|
|
Analogs
-
3871393
-
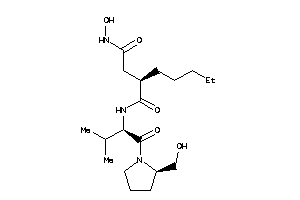
-
3871392
-
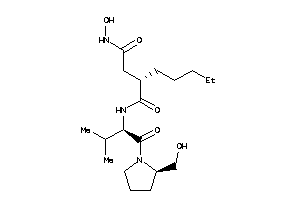
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DEF-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
5 |
0.43 |
Binding ≤ 10μM
|
|
Q9JN24-1-B |
Peptide Deformylase (cluster #1 Of 1), Bacterial |
Bacteria |
140 |
0.36 |
Binding ≤ 10μM |
|
DEF1A-1-E |
Peptide Deformylase 1A, Chloroplastic (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.39 |
Binding ≤ 10μM
|
|
DEFM-1-E |
Peptide Deformylase Mitochondrial (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.32 |
Binding ≤ 10μM
|
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.28 |
Binding ≤ 10μM
|
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM |
|
MMP1-2-E |
Matrix Metalloproteinase-1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.31 |
Binding ≤ 10μM
|
|
MMP2-1-E |
72 KDa Type IV Collagenase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.29 |
Binding ≤ 10μM
|
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.30 |
Binding ≤ 10μM |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.27 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.35 |
Binding ≤ 10μM |
|
MMP9-2-E |
Matrix Metalloproteinase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
330 |
0.34 |
Binding ≤ 10μM |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.26 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2512 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
2.32 |
-16.44 |
4 |
8 |
0 |
119 |
385.505 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.51 |
3.4 |
-55.95 |
3 |
8 |
-1 |
122 |
384.497 |
11 |
↓
|
|
|
|
Analogs
-
4899476
-
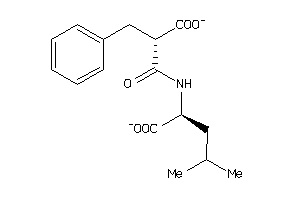
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
0.1 |
-123.46 |
1 |
6 |
-2 |
109 |
305.33 |
8 |
↓
|
|
|
|
Analogs
-
2391109
-
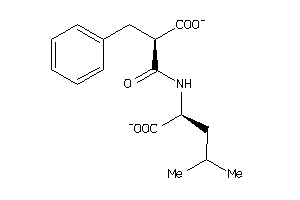
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.27 |
0.54 |
-106.5 |
1 |
6 |
-2 |
109 |
305.33 |
8 |
↓
|
|
|
|
Analogs
-
44625037
-
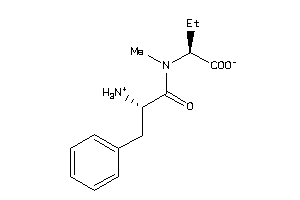
-
44625040
-
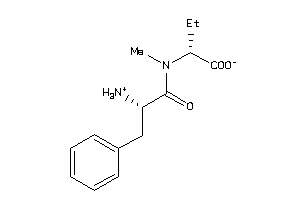
-
44625115
-
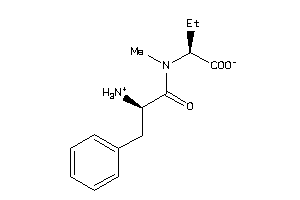
-
44625118
-
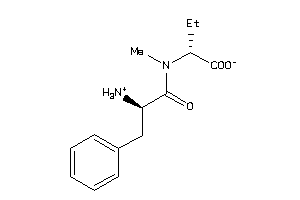
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.79 |
-3.43 |
-60.11 |
4 |
5 |
0 |
96 |
236.271 |
5 |
↓
|
|
|
|
Analogs
-
44625037
-

-
44625040
-

-
44625115
-

-
44625118
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.79 |
-3.43 |
-60.89 |
4 |
5 |
0 |
96 |
236.271 |
5 |
↓
|
|
|
|
Analogs
-
44625037
-

-
44625040
-

-
44625115
-

-
44625118
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
|
S15A1-1-E |
Oligopeptide Transporter Small Intestine Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.79 |
-3.43 |
-60.91 |
4 |
5 |
0 |
96 |
236.271 |
5 |
↓
|
|
|
|
Analogs
-
44625037
-

-
44625040
-

-
44625115
-

-
44625118
-

Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.79 |
-3.46 |
-60.18 |
4 |
5 |
0 |
96 |
236.271 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.43 |
Binding ≤ 10μM
|
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.43 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.39 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
63 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
-1.08 |
-67.14 |
1 |
6 |
-1 |
89 |
377.486 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.92 |
-2.03 |
-116.66 |
1 |
6 |
-2 |
89 |
376.478 |
6 |
↓
|
|