|
|
Analogs
-
4228257
-
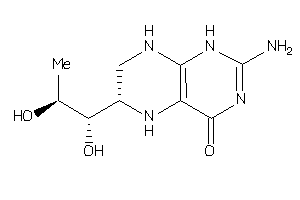
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1120 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.44 |
-11.69 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-1.15 |
-5.1 |
-20.07 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
300 |
0.34 |
Binding ≤ 10μM |
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
91 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
9.12 |
-18.56 |
1 |
6 |
0 |
76 |
367.359 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.02 |
7.17 |
-56.87 |
0 |
6 |
-1 |
80 |
366.351 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
80 |
0.50 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
90 |
0.49 |
Binding ≤ 10μM
|
|
NOS3-3-E |
Nitric Oxide Synthase, Endothelial (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
8.38 |
-39.13 |
1 |
5 |
1 |
53 |
298.794 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.73 |
8.03 |
-35.47 |
1 |
5 |
1 |
53 |
298.794 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
120 |
0.39 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
NOS3-3-E |
Nitric Oxide Synthase, Endothelial (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
3500 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
9.82 |
-28.79 |
4 |
3 |
1 |
50 |
370.929 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.18 |
9.36 |
-7.3 |
3 |
3 |
0 |
50 |
369.921 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.31 |
11.22 |
-52.4 |
4 |
3 |
1 |
52 |
370.929 |
8 |
↓
|
|
|
|
Analogs
-
6709
-
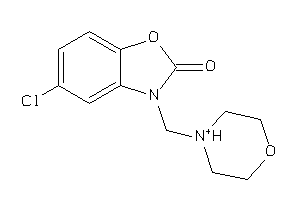
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
5600 |
0.41 |
Binding ≤ 10μM |
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
800 |
0.47 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.76 |
4.63 |
-8.47 |
0 |
5 |
0 |
48 |
268.7 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4400 |
0.42 |
Binding ≤ 10μM |
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
3500 |
0.42 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
8.67 |
-42 |
1 |
4 |
1 |
40 |
285.776 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
6.32 |
-8.81 |
0 |
4 |
0 |
38 |
284.768 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
75 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
8.84 |
-74.72 |
4 |
4 |
2 |
54 |
301.434 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.45 |
6.27 |
-7.86 |
2 |
4 |
0 |
51 |
299.418 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.45 |
8.74 |
-44.77 |
3 |
4 |
1 |
53 |
300.426 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
100 |
0.35 |
Binding ≤ 10μM
|
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
840 |
0.30 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
900 |
0.30 |
Binding ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
900 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.10 |
-2.7 |
-41.18 |
4 |
7 |
1 |
109 |
383.382 |
1 |
↓
|
|
|
|
Analogs
-
39246631
-
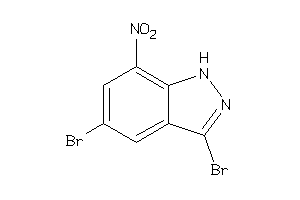
Draw
Identity
99%
90%
80%
70%
Vendors
And 21 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1800 |
0.62 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
290 |
0.70 |
Binding ≤ 10μM
|
|
NOS3-2-E |
Nitric Oxide Synthase, Endothelial (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2000 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
4.41 |
-8.35 |
1 |
5 |
0 |
75 |
242.032 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.47 |
4.28 |
-39.17 |
0 |
5 |
-1 |
73 |
241.024 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2400 |
0.66 |
Binding ≤ 10μM
|
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4000 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
4.87 |
-24.63 |
3 |
2 |
1 |
40 |
163.244 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4600 |
0.42 |
Binding ≤ 10μM |
|
NOS2-4-E |
Nitric Oxide Synthase, Inducible (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
1500 |
0.45 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
1.48 |
-38.48 |
1 |
4 |
1 |
39 |
267.736 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
3.43 |
-33.02 |
3 |
3 |
1 |
47 |
179.243 |
4 |
↓
|
|
|
|
Analogs
-
22172988
-
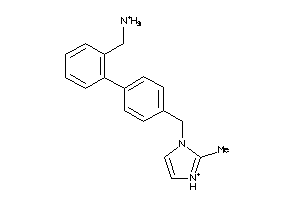
-
22173678
-
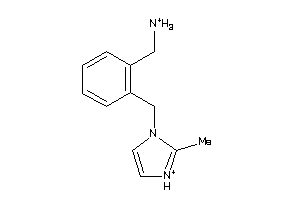
-
36373992
-
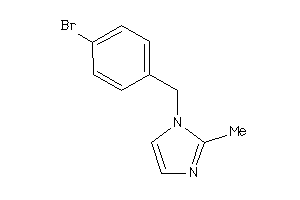
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
7.55 |
-7.81 |
0 |
2 |
0 |
18 |
172.231 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.68 |
8.02 |
-29.34 |
1 |
2 |
1 |
19 |
173.239 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6300 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
2.43 |
-6.87 |
1 |
3 |
0 |
38 |
148.165 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
850 |
0.35 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
0.58 |
-22.96 |
1 |
4 |
0 |
59 |
323.392 |
2 |
↓
|
|