|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
126 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
1.36 |
-10.41 |
0 |
4 |
0 |
42 |
421.711 |
4 |
↓
|
|
|
|
Analogs
-
6941598
-
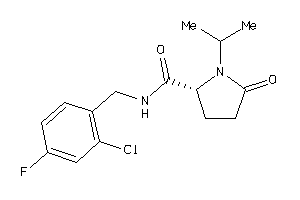
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
316 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
6.77 |
-14.47 |
1 |
4 |
0 |
49 |
312.772 |
4 |
↓
|
|
|
|
Analogs
-
6941592
-
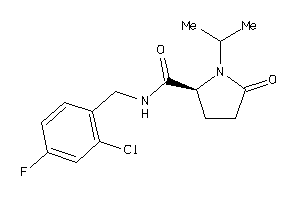
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
316 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
6.26 |
-13.33 |
1 |
4 |
0 |
49 |
312.772 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
92 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
12.83 |
-42.14 |
2 |
9 |
-1 |
122 |
473.557 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
3.76 |
13.91 |
-14.98 |
3 |
9 |
0 |
121 |
474.565 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
3.76 |
12.03 |
-17.11 |
3 |
9 |
0 |
121 |
474.565 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
20 |
0.31 |
Binding ≤ 10μM
|
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
92 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
12.82 |
-44 |
2 |
9 |
-1 |
122 |
473.557 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
3.76 |
14.27 |
-16.14 |
3 |
9 |
0 |
121 |
474.565 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
3.76 |
12.04 |
-18.02 |
3 |
9 |
0 |
121 |
474.565 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
316 |
0.46 |
Binding ≤ 10μM
|
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
126 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
6.91 |
-15.08 |
0 |
5 |
0 |
57 |
306.156 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
11.77 |
-35.76 |
1 |
5 |
-1 |
75 |
314.372 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
3.48 |
13.82 |
-12.17 |
2 |
5 |
0 |
73 |
315.38 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.66 |
13.67 |
-31.74 |
1 |
5 |
-1 |
78 |
314.372 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.72 |
0.39 |
-15.28 |
1 |
4 |
0 |
46 |
319.408 |
5 |
↓
|
|
|
|
Analogs
-
20887150
-
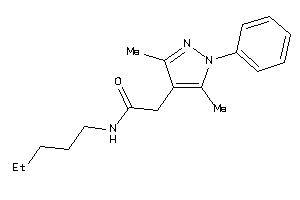
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX7-1-E |
P2X Purinoceptor 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
158 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
0.15 |
-15.51 |
1 |
4 |
0 |
46 |
325.456 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
-9.06 |
-32.14 |
0 |
11 |
0 |
130 |
721.861 |
10 |
↓
|
|