|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8128 |
0.40 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8100 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.82 |
4.37 |
-112.3 |
1 |
6 |
-2 |
109 |
267.262 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.41 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9400 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
1.97 |
-124.27 |
3 |
7 |
-1 |
126 |
269.258 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUS3-1-E |
Dual Specificity Protein Phosphatase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.22 |
Binding ≤ 10μM
|
|
PP1A-2-E |
Serine/threonine Protein Phosphatase PP1-alpha Catalytic Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8100 |
0.21 |
Binding ≤ 10μM
|
|
PP1G-2-E |
Serine/threonine Protein Phosphatase PP1-gamma Catalytic Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8100 |
0.21 |
Binding ≤ 10μM
|
|
PPAC-1-E |
Low Molecular Weight Phosphotyrosine Protein Phosphatase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.22 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9900 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.75 |
0.27 |
-98.84 |
3 |
12 |
-2 |
188 |
510.55 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.75 |
0.34 |
-185.53 |
2 |
12 |
-3 |
190 |
509.542 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.75 |
0.34 |
-189.06 |
2 |
12 |
-3 |
190 |
509.542 |
8 |
↓
|
|
|
|
Analogs
-
2168323
-
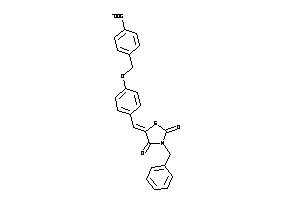
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPAC-1-E |
Low Molecular Weight Phosphotyrosine Protein Phosphatase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5600 |
0.23 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1600 |
0.25 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1300 |
0.26 |
Binding ≤ 10μM
|
|
PTPRB-1-E |
Receptor-type Tyrosine-protein Phosphatase Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
400 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.71 |
12.08 |
-55.76 |
0 |
6 |
-1 |
88 |
444.488 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
35 |
0.29 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
35 |
0.29 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
3 |
-58.35 |
2 |
9 |
-1 |
144 |
523.616 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.12 |
3.19 |
-78.67 |
3 |
9 |
0 |
146 |
524.624 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.32 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.19 |
10.46 |
-131.2 |
1 |
9 |
-2 |
139 |
607.504 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.38 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9200 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
7.49 |
-127.92 |
0 |
5 |
-2 |
89 |
306.32 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
140 |
0.27 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
4.39 |
-20.52 |
3 |
10 |
0 |
141 |
525.612 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3200 |
0.23 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7000 |
0.21 |
Binding ≤ 10μM
|
|
PTPRC-1-E |
Leukocyte Common Antigen (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9700 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
1.39 |
-62.88 |
1 |
6 |
-1 |
95 |
470.526 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.28 |
Binding ≤ 10μM
|
|
PTN2-1-E |
T-cell Protein-tyrosine Phosphatase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5300 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
10.46 |
-43.8 |
0 |
7 |
-1 |
100 |
421.863 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.49 |
9.46 |
-12.69 |
1 |
7 |
0 |
97 |
422.871 |
4 |
↓
|
|