|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
-1.26 |
-7.04 |
2 |
3 |
0 |
49 |
134.138 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.53 |
0.53 |
-47.28 |
1 |
3 |
-1 |
52 |
133.13 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
5.37 |
-39.81 |
1 |
4 |
-1 |
73 |
222.607 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
7600 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
8.42 |
-7.84 |
2 |
4 |
0 |
58 |
330.185 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
1940 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.69 |
-11.41 |
1 |
3 |
0 |
38 |
238.29 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
5250 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
-2.47 |
-10.02 |
2 |
3 |
0 |
48 |
210.236 |
1 |
↓
|
|
|
|
Analogs
-
3880991
-
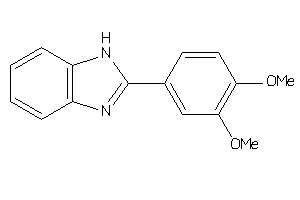
-
34575181
-
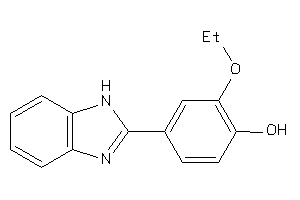
-
42173246
-
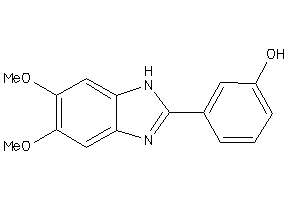
-
42173274
-
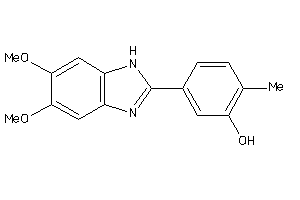
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
1190 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
-2.1 |
-11.32 |
2 |
4 |
0 |
58 |
240.262 |
2 |
↓
|
|
|
|
Analogs
-
3651042
-
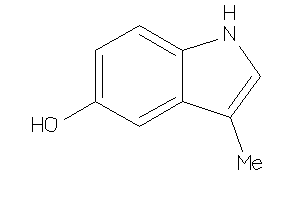
Draw
Identity
99%
90%
80%
70%
Vendors
And 69 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-3-E |
Serotonin 1a (5-HT1a) Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
5HT1E-1-E |
Serotonin 1e (5-HT1e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.90 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
5HT2A-2-E |
Serotonin 2a (5-HT2a) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.66 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
680 |
0.66 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
680 |
0.66 |
Binding ≤ 10μM
|
|
5HT3A-4-E |
Serotonin 3a (5-HT3a) Receptor (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.76 |
Binding ≤ 10μM |
|
5HT3C-1-E |
Serotonin 3c (5-HT3c) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT3D-1-E |
Serotonin 3d (5-HT3d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT3E-1-E |
Serotonin 3e (5-HT3e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
120 |
0.75 |
Binding ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
37 |
0.80 |
Binding ≤ 10μM |
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
93 |
0.76 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.87 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
79 |
0.76 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9690 |
0.54 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
523 |
0.68 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
740 |
0.66 |
Binding ≤ 10μM
|
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.86 |
Functional ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Functional ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Functional ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.87 |
Functional ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.77 |
Functional ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Functional ≤ 10μM
|
|
5HT2C-1-E |
5-hydroxytryptamine Receptor 2C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Functional ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.68 |
Functional ≤ 10μM
|
|
5HT4R-1-E |
Serotonin 4 (5-HT4) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.90 |
Functional ≤ 10μM |
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.84 |
Functional ≤ 10μM
|
|
DRD2-1-E |
Dopamine D2 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50588-4-O |
Canis Familiaris (cluster #4 Of 7), Other |
Other |
210 |
0.72 |
Functional ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 8), Other |
Other |
300 |
0.70 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
919 |
0.65 |
Functional ≤ 10μM
|
|
Z80334-1-O |
NG108-15 (Neuroblastoma-glioma Hybrid Cells) (cluster #1 Of 1), Other |
Other |
298 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
0.5 |
-47.88 |
5 |
3 |
1 |
64 |
177.227 |
2 |
↓
|
|
|
|
Analogs
-
8548768
-
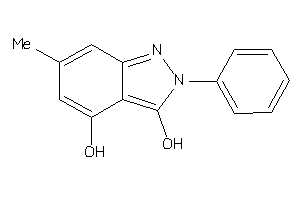
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
4.76 |
-10.42 |
1 |
3 |
0 |
38 |
210.236 |
1 |
↓
|
|
|
|
Analogs
-
34876312
-
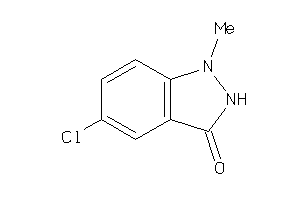
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX5-4-E |
Arachidonate 5-lipoxygenase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
3000 |
0.70 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
3000 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
2.66 |
-11.18 |
1 |
3 |
0 |
38 |
148.165 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
4.46 |
-39.29 |
3 |
3 |
1 |
40 |
205.281 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
8.9 |
-7.63 |
2 |
4 |
0 |
58 |
377.185 |
3 |
↓
|
|
|
|
Analogs
-
34354758
-
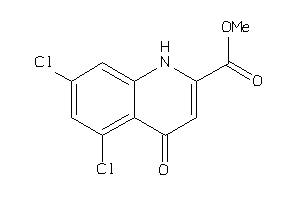
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GLRA1-1-E |
Glycine Receptor Subunit Alpha-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRA2-1-E |
Glycine Receptor Subunit Alpha-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRA3-1-E |
Glycine Receptor Alpha-3 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GLRB-1-E |
Glycine Receptor Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.62 |
Binding ≤ 10μM
|
|
GRIA1-2-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA2-1-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA3-1-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
GRIA4-1-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
500 |
0.55 |
Binding ≤ 10μM |
|
NMD3A-5-E |
Glutamate [NMDA] Receptor Subunit 3A (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-5-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE1-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE2-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE3-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDE4-5-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMDZ1-3-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
68 |
0.63 |
Binding ≤ 10μM |
|
NMD3B-1-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Functional ≤ 10μM
|
|
Z104302-4-O |
Glutamate NMDA Receptor (cluster #4 Of 7), Other |
Other |
90 |
0.62 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.79 |
5.87 |
-38.83 |
1 |
4 |
-1 |
73 |
257.052 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
5.21 |
-12.17 |
1 |
4 |
0 |
51 |
225.251 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
1.96 |
3.26 |
-11.44 |
1 |
4 |
0 |
51 |
225.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
3.06 |
-11.19 |
1 |
3 |
0 |
38 |
148.165 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.60 |
1.13 |
-11.88 |
1 |
3 |
0 |
38 |
148.165 |
0 |
↓
|
|
|
|
Analogs
-
6117549
-
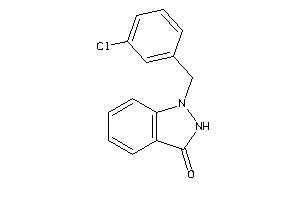
Draw
Identity
99%
90%
80%
70%
Vendors
And 38 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX5-4-E |
Arachidonate 5-lipoxygenase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
3400 |
0.45 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
3400 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
3.9 |
-8.64 |
1 |
3 |
0 |
38 |
224.263 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
2.74 |
6.51 |
-11.9 |
1 |
3 |
0 |
38 |
224.263 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.19 |
4.66 |
-55.96 |
0 |
3 |
-1 |
41 |
223.255 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
3200 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.64 |
8.32 |
-7.93 |
2 |
4 |
0 |
58 |
285.734 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
-
- ZP-BB-AS004416
- ZP-BB-AS004424
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
7.21 |
-31.75 |
2 |
3 |
1 |
29 |
212.276 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.40 |
-0.5 |
-37.56 |
2 |
3 |
1 |
31 |
210.26 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
-0.25 |
-17.95 |
1 |
5 |
0 |
60 |
294.31 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
60 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
6.06 |
-43.37 |
3 |
2 |
1 |
32 |
201.293 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
200 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
-0.13 |
-15.2 |
1 |
6 |
0 |
69 |
324.336 |
5 |
↓
|
|
|
|
Analogs
-
23382757
-
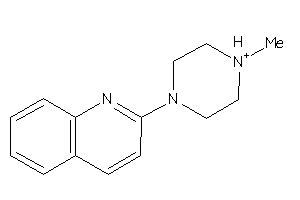
-
37037830
-
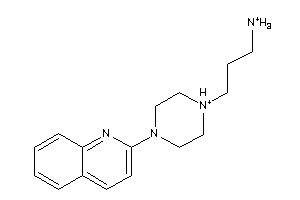
-
37039478
-
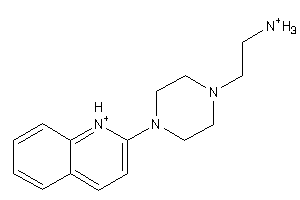
-
41677432
-
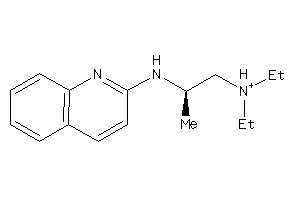
-
41677434
-
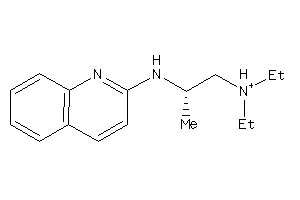
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3600 |
0.48 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7079 |
0.45 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3715 |
0.48 |
Binding ≤ 10μM
|
|
5HT1E-1-E |
Serotonin 1e (5-HT1e) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.58 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
230 |
0.58 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3467 |
0.48 |
Binding ≤ 10μM
|
|
5HT2B-2-E |
Serotonin 2b (5-HT2b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
645 |
0.54 |
Binding ≤ 10μM
|
|
5HT2C-2-E |
Serotonin 2c (5-HT2c) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
62 |
0.63 |
Binding ≤ 10μM
|
|
5HT3A-2-E |
Serotonin 3a (5-HT3a) Receptor (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.73 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6 |
0.72 |
Binding ≤ 10μM
|
|
5HT3C-2-E |
Serotonin 3c (5-HT3c) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.76 |
Binding ≤ 10μM
|
|
5HT3D-2-E |
Serotonin 3d (5-HT3d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.76 |
Binding ≤ 10μM
|
|
5HT3E-2-E |
Serotonin 3e (5-HT3e) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.76 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3600 |
0.48 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3033 |
0.48 |
Binding ≤ 10μM
|
|
SC6A4-2-E |
Serotonin Transporter (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
63 |
0.63 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.79 |
Functional ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.79 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.45 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
204 |
0.59 |
Functional ≤ 10μM
|
|
Z80334-1-O |
NG108-15 (Neuroblastoma-glioma Hybrid Cells) (cluster #1 Of 1), Other |
Other |
25 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.68 |
6.11 |
-45.53 |
2 |
3 |
1 |
33 |
214.292 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.68 |
4.81 |
-7.58 |
1 |
3 |
0 |
28 |
213.284 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 32 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
5150 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.28 |
4.61 |
-10.37 |
2 |
3 |
0 |
49 |
210.236 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
7700 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
-0.1 |
-11.82 |
1 |
3 |
0 |
37 |
224.263 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
4829003
-
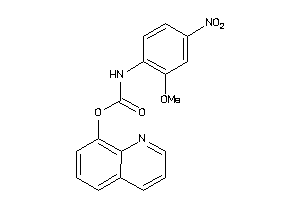
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
7000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
-0.12 |
-15.45 |
1 |
5 |
0 |
60 |
294.31 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX5-4-E |
Arachidonate 5-lipoxygenase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
360 |
0.53 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
360 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
7.01 |
-11.97 |
1 |
3 |
0 |
38 |
224.263 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.19 |
5.03 |
-12.05 |
1 |
3 |
0 |
38 |
224.263 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX5-4-E |
Arachidonate 5-lipoxygenase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
1800 |
0.50 |
Functional ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
1800 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
4.29 |
-12.93 |
1 |
4 |
0 |
51 |
231.28 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.36 |
2.48 |
-13.84 |
1 |
4 |
0 |
51 |
231.28 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 84 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PERM-1-E |
Myeloperoxidase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
940 |
0.65 |
Binding ≤ 10μM
|
|
Z50597-10-O |
Rattus Norvegicus (cluster #10 Of 12), Other |
Other |
130 |
0.74 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.24 |
3.43 |
-47.2 |
4 |
2 |
1 |
43 |
179.218 |
2 |
↓
|
|