|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NFKB1-1-E |
Nuclear Factor NF-kappa-B P105 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.56 |
Binding ≤ 10μM
|
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
900 |
0.60 |
Functional ≤ 10μM
|
|
Z80164-6-O |
HT-1080 (Fibrosarcoma Cells) (cluster #6 Of 6), Other |
Other |
2600 |
0.56 |
Functional ≤ 10μM
|
|
Z80186-5-O |
K562 (Erythroleukemia Cells) (cluster #5 Of 11), Other |
Other |
1200 |
0.59 |
Functional ≤ 10μM |
|
Z80211-5-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #5 Of 5), Other |
Other |
900 |
0.60 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
4400 |
0.54 |
Functional ≤ 10μM |
|
Z81072-10-O |
Jurkat (Acute Leukemic T-cells) (cluster #10 Of 10), Other |
Other |
900 |
0.60 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
2.86 |
-14.35 |
0 |
3 |
0 |
43 |
186.166 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 34 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
9500 |
0.50 |
Functional ≤ 10μM
|
|
Z80211-5-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #5 Of 5), Other |
Other |
7300 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
3.16 |
-12.54 |
1 |
4 |
0 |
60 |
192.17 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
5870 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
6.19 |
-13.38 |
1 |
8 |
0 |
97 |
388.372 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
8100 |
0.31 |
Binding ≤ 10μM
|
|
MTOR-1-E |
Serine/threonine-protein Kinase MTOR (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.32 |
Binding ≤ 10μM
|
|
P3C2B-1-E |
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.35 |
Binding ≤ 10μM
|
|
P85A-2-E |
PI3-kinase P85-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Binding ≤ 10μM
|
|
PIM1-1-E |
Serine/threonine-protein Kinase PIM1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
820 |
0.37 |
Binding ≤ 10μM
|
|
PIM3-1-E |
Serine/threonine-protein Kinase PIM3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.36 |
Binding ≤ 10μM
|
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
PK3CB-1-E |
PI3-kinase P110-beta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.37 |
Binding ≤ 10μM
|
|
PK3CD-1-E |
PI3-kinase P110-delta Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
PK3CG-1-E |
PI3-kinase P110-gamma Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7260 |
0.31 |
Binding ≤ 10μM
|
|
PLK1-1-E |
Serine/threonine-protein Kinase PLK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2100 |
0.35 |
Binding ≤ 10μM
|
|
PLK3-1-E |
Serine/threonine-protein Kinase PLK3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.34 |
Binding ≤ 10μM
|
|
PRKDC-1-E |
DNA-dependent Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.32 |
Binding ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
8400 |
0.31 |
Functional ≤ 10μM
|
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
9940 |
0.30 |
Functional ≤ 10μM |
|
Z80418-2-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #2 Of 9), Other |
Other |
10000 |
0.30 |
Functional ≤ 10μM
|
|
Z80548-3-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #3 Of 5), Other |
Other |
1650 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
8400 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.05 |
6.42 |
-28.01 |
1 |
4 |
1 |
46 |
308.357 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.71 |
5.27 |
-99.28 |
7 |
7 |
2 |
113 |
408.481 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.64 |
8.22 |
-10.99 |
0 |
4 |
0 |
43 |
307.349 |
2 |
↓
|
|
|
|
Analogs
-
5999141
-
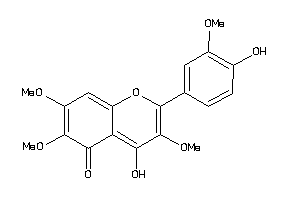
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
7260 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
5.77 |
-13.78 |
1 |
8 |
0 |
97 |
388.372 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.90 |
6.84 |
-59.48 |
0 |
8 |
-1 |
99 |
387.364 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 33 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
3350 |
0.48 |
Functional ≤ 10μM
|
|
Z80164-6-O |
HT-1080 (Fibrosarcoma Cells) (cluster #6 Of 6), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
5.76 |
-10.08 |
0 |
4 |
0 |
53 |
216.192 |
1 |
↓
|
|
|
|
Analogs
-
5430812
-
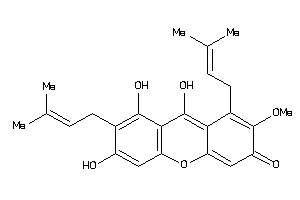
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
4.64 |
-11.98 |
4 |
6 |
0 |
111 |
396.439 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.05 |
6.27 |
-51.41 |
3 |
6 |
-1 |
114 |
395.431 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.05 |
6.19 |
-66.49 |
3 |
6 |
-1 |
114 |
395.431 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NFKB1-1-E |
Nuclear Factor NF-kappa-B P105 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.23 |
Binding ≤ 10μM
|
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
7600 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.39 |
1.56 |
-10.85 |
2 |
6 |
0 |
89 |
424.493 |
6 |
↓
|
|
|
|
Analogs
-
5195818
-
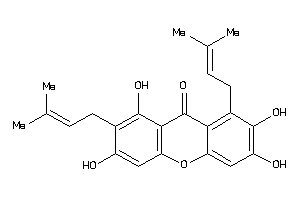
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.32 |
6.65 |
-10.92 |
3 |
6 |
0 |
100 |
410.466 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
5.42 |
7.43 |
-44.09 |
2 |
6 |
-1 |
103 |
409.458 |
5 |
↓
|
|
|
|
Analogs
-
39291087
-
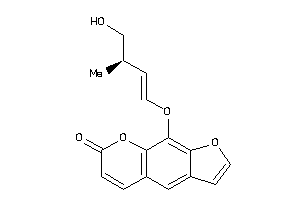
-
39291089
-
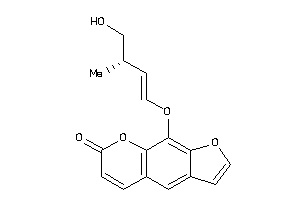
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
6.1 |
-16.38 |
0 |
4 |
0 |
53 |
216.192 |
1 |
↓
|
|