|
|
Analogs
-
3647804
-
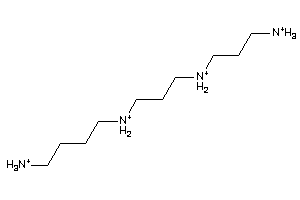
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-8-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #8 Of 12), Other |
Other |
300 |
0.51 |
Functional ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
500 |
0.49 |
Functional ≤ 10μM
|
|
Z80878-2-O |
NCI-H157 (Non-small Cell Lung Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
2900 |
0.43 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.12 |
5.6 |
-259.45 |
8 |
4 |
4 |
66 |
262.486 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.11 |
4.24 |
-175.45 |
7 |
4 |
3 |
62 |
261.478 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.11 |
2.89 |
-84.16 |
6 |
4 |
2 |
57 |
260.47 |
15 |
↓
|
|
|
|
Analogs
-
1532612
-
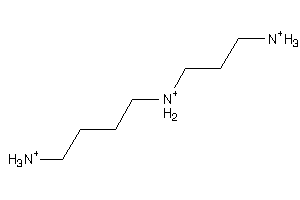
Draw
Identity
99%
90%
80%
70%
Vendors
And 36 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH14-2-E |
Carbonic Anhydrase XIV (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
860 |
0.61 |
Binding ≤ 10μM
|
|
CAH4-5-E |
Carbonic Anhydrase IV (cluster #5 Of 16), Eukaryotic |
Eukaryotes |
10 |
0.80 |
Binding ≤ 10μM
|
|
CAH5A-4-E |
Carbonic Anhydrase VA (cluster #4 Of 10), Eukaryotic |
Eukaryotes |
840 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B-5-E |
Carbonic Anhydrase VB (cluster #5 Of 9), Eukaryotic |
Eukaryotes |
830 |
0.61 |
Binding ≤ 10μM
|
|
CAH6-3-E |
Carbonic Anhydrase VI (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
990 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-3-E |
Carbonic Anhydrase VII (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
710 |
0.61 |
Binding ≤ 10μM
|
|
CASP2-3-E |
Caspase-2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.51 |
Binding ≤ 10μM
|
|
Z100250-2-O |
Calf Thymus DNA (cluster #2 Of 2), Other |
Other |
4300 |
0.54 |
Binding ≤ 10μM
|
|
Z104302-6-O |
Glutamate NMDA Receptor (cluster #6 Of 7), Other |
Other |
5200 |
0.53 |
Binding ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
390 |
0.64 |
Functional ≤ 10μM
|
|
Z80608-2-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #2 Of 4), Other |
Other |
200 |
0.67 |
Functional ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
920 |
0.60 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
280 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.13 |
0.14 |
-262.68 |
10 |
4 |
4 |
88 |
206.378 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-0.62 |
-97.52 |
8 |
4 |
2 |
85 |
204.362 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.13 |
-1.59 |
-88.23 |
8 |
4 |
2 |
82 |
204.362 |
11 |
↓
|
|
|
|
Analogs
-
1655626
-
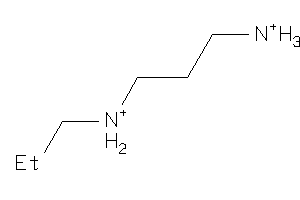
-
19911753
-
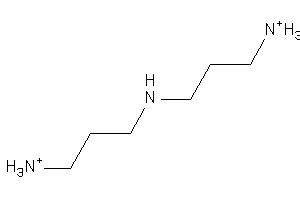
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
900 |
0.94 |
Functional ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
9500 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.10 |
-6.75 |
-183.75 |
8 |
3 |
3 |
71 |
134.247 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
3000 |
0.70 |
Functional ≤ 10μM
|
|
Z80712-6-O |
T47D (Breast Carcinoma Cells) (cluster #6 Of 7), Other |
Other |
2700 |
0.71 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.56 |
-6.5 |
-167.66 |
8 |
3 |
3 |
71 |
162.301 |
8 |
↓
|
|
|
|
Analogs
-
5178755
-
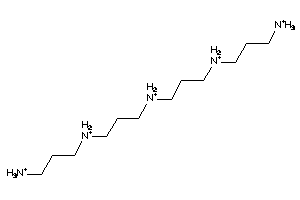
-
5700567
-
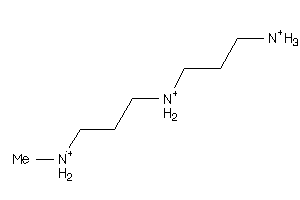
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.38 |
4.81 |
-271.51 |
8 |
4 |
4 |
66 |
248.459 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80019-3-O |
A-427 (Lung Carcinoma Cells) (cluster #3 Of 4), Other |
Other |
8000 |
0.59 |
Functional ≤ 10μM
|
|
Z80088-3-O |
CHO (Ovarian Cells) (cluster #3 Of 4), Other |
Other |
8200 |
0.59 |
Functional ≤ 10μM
|
|
Z80186-11-O |
K562 (Erythroleukemia Cells) (cluster #11 Of 11), Other |
Other |
9000 |
0.59 |
Functional ≤ 10μM
|
|
Z80193-7-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #7 Of 12), Other |
Other |
4500 |
0.62 |
Functional ≤ 10μM
|
|
Z80224-13-O |
MCF7 (Breast Carcinoma Cells) (cluster #13 Of 14), Other |
Other |
4500 |
0.62 |
Functional ≤ 10μM
|
|
Z80362-3-O |
P388 (Lymphoma Cells) (cluster #3 Of 8), Other |
Other |
5900 |
0.61 |
Functional ≤ 10μM
|
|
Z80565-2-O |
U-87 MG (Glioblastoma Cells) (cluster #2 Of 2), Other |
Other |
1420 |
0.68 |
Functional ≤ 10μM
|
|
Z80742-3-O |
C6 (Glioma Cells) (cluster #3 Of 3), Other |
Other |
9300 |
0.59 |
Functional ≤ 10μM
|
|
Z80876-1-O |
NCI-H125 (cluster #1 Of 1), Other |
Other |
9000 |
0.59 |
Functional ≤ 10μM
|
|
Z81184-4-O |
LOX IMVI (Melanoma Cells) (cluster #4 Of 5), Other |
Other |
7000 |
0.60 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
7400 |
0.60 |
Functional ≤ 10μM
|
|
Z81281-5-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #5 Of 5), Other |
Other |
5000 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
2.01 |
-5.18 |
1 |
5 |
0 |
62 |
214.052 |
5 |
↓
|
|
|
|
Analogs
-
5947366
-
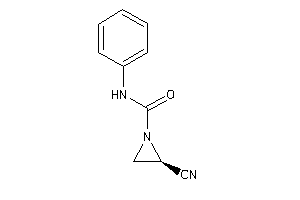
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
3.38 |
-17.44 |
1 |
4 |
0 |
56 |
187.202 |
1 |
↓
|
|
|
|
Analogs
-
3916314
-
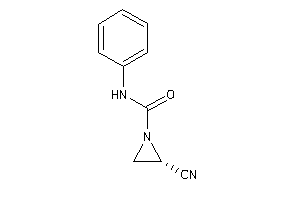
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
3.38 |
-17.47 |
1 |
4 |
0 |
56 |
187.202 |
1 |
↓
|
|