|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.65 |
-4.34 |
-17.19 |
6 |
8 |
0 |
137 |
239.235 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.07 |
-4.43 |
-43.98 |
7 |
8 |
1 |
138 |
240.243 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.07 |
-4.17 |
-38.96 |
7 |
8 |
1 |
138 |
240.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
1.41 |
-7.7 |
4 |
5 |
0 |
90 |
213.244 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.49 |
1.89 |
-34.29 |
5 |
5 |
1 |
91 |
214.252 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.49 |
3.21 |
-33.91 |
5 |
5 |
1 |
91 |
214.252 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
-0.34 |
-1.87 |
-50.91 |
3 |
4 |
-1 |
82 |
125.107 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
4.71 |
-38.4 |
4 |
6 |
1 |
84 |
198.25 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.19 |
4.32 |
-13.13 |
3 |
6 |
0 |
83 |
197.242 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.23 |
Binding ≤ 10μM
|
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.23 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.32 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3190 |
0.25 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_MOUSE |
P05132
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Mouse |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB_MOUSE |
P68181
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Mouse |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAP2_BOVIN |
P00515
|
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit, Bovin |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
1500 |
0.26 |
Binding ≤ 10μM
|
|
P2RX1_HUMAN |
P51575
|
P2X Purinoceptor 1, Human |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2_RAT |
P49653
|
P2X Purinoceptor 2, Rat |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3_HUMAN |
P56373
|
P2X Purinoceptor 3, Human |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4_RAT |
P51577
|
P2X Purinoceptor 4, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX4_HUMAN |
Q99571
|
P2X Purinoceptor 4, Human |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5_RAT |
P51578
|
P2X Purinoceptor 5, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6_RAT |
P51579
|
P2X Purinoceptor 6, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
14 |
0.35 |
Functional ≤ 10μM
|
|
P2Y11_HUMAN |
Q96G91
|
Purinergic Receptor P2Y11, Human |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12_RAT |
Q9EPX4
|
Purinergic Receptor P2Y12, Rat |
3190 |
0.25 |
Functional ≤ 10μM
|
|
P2RY2_HUMAN |
P41231
|
Purinergic Receptor P2Y2, Human |
230 |
0.30 |
Functional ≤ 10μM
|
|
P2RY4_RAT |
O35811
|
Pyrimidinergic Receptor P2Y4, Rat |
1800 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
GRP78-1-E |
78 KDa Glucose-regulated Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3830 |
0.28 |
Binding ≤ 10μM
|
|
HSP7C-1-E |
Heat Shock Cognate 71 KDa Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
69 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
0.91 |
0.47 |
Binding ≤ 1μM
|
|
HSP7C_BOVIN |
P19120
|
Heat Shock Cognate 71 KDa Protein, Bovin |
110 |
0.36 |
Binding ≤ 1μM
|
|
HSP7C_HUMAN |
P11142
|
Heat Shock Cognate 71 KDa Protein, Human |
260 |
0.34 |
Binding ≤ 1μM
|
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
0.91 |
0.47 |
Binding ≤ 10μM
|
|
GRP78_HUMAN |
P11021
|
78 KDa Glucose-regulated Protein, Human |
2170 |
0.29 |
Binding ≤ 10μM
|
|
KAPCA_MOUSE |
P05132
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Mouse |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB_MOUSE |
P68181
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Mouse |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAP2_BOVIN |
P00515
|
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit, Bovin |
10000 |
0.26 |
Binding ≤ 10μM
|
|
HSP7C_BOVIN |
P19120
|
Heat Shock Cognate 71 KDa Protein, Bovin |
110 |
0.36 |
Binding ≤ 10μM
|
|
HSP7C_HUMAN |
P11142
|
Heat Shock Cognate 71 KDa Protein, Human |
260 |
0.34 |
Binding ≤ 10μM
|
|
P2RX1_HUMAN |
P51575
|
P2X Purinoceptor 1, Human |
10000 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1_MELGA |
P49652
|
P2Y Purinoceptor 1, Melga |
100 |
0.36 |
Functional ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
14 |
0.41 |
Functional ≤ 10μM
|
|
P2Y12_RAT |
Q9EPX4
|
Purinergic Receptor P2Y12, Rat |
69 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
3.79 |
9.28 |
-36.43 |
1 |
2 |
1 |
8 |
233.379 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
8215755
-
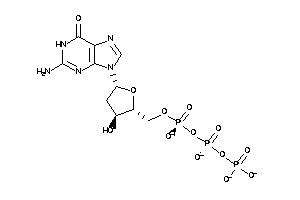
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.25 |
-5.62 |
-178.53 |
5 |
18 |
-3 |
288 |
504.158 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.55 |
-2.36 |
-236.83 |
5 |
18 |
-3 |
287 |
504.158 |
8 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.72 |
-3.32 |
-50.87 |
3 |
7 |
-1 |
121 |
178.131 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.72 |
-3.23 |
-47.68 |
3 |
7 |
-1 |
121 |
178.131 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.26 |
-5.46 |
-104.45 |
2 |
7 |
-2 |
124 |
177.123 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.12 |
-0.82 |
-55.23 |
5 |
6 |
1 |
102 |
152.137 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.34 |
-3.23 |
-62.94 |
3 |
6 |
-1 |
104 |
150.121 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.34 |
-2.46 |
-35.05 |
3 |
6 |
-1 |
104 |
150.121 |
0 |
↓
|
|
|
|
Analogs
-
8615332
-
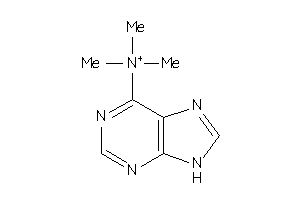
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
5.39 |
-9.94 |
1 |
5 |
0 |
58 |
163.184 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.85 |
4.12 |
-31.1 |
2 |
5 |
1 |
59 |
164.192 |
1 |
↓
|
|
|
|
Analogs
-
32141845
-
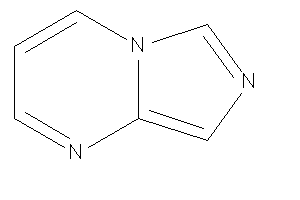
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.01 |
5.53 |
-30.01 |
1 |
5 |
0 |
59 |
159.152 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.01 |
5.6 |
-15.57 |
1 |
5 |
0 |
59 |
159.152 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
-0.01 |
5.8 |
-18.06 |
1 |
5 |
0 |
59 |
159.152 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GUAD-1-E |
Guanine Deaminase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4340 |
0.63 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.99 |
-5.59 |
-30.42 |
3 |
7 |
-1 |
117 |
167.104 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.44 |
-3.48 |
-11.55 |
4 |
7 |
0 |
114 |
168.112 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.83 |
-3.4 |
-55.25 |
4 |
7 |
-1 |
124 |
166.12 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.66 |
-6.13 |
-90.31 |
1 |
7 |
-2 |
122 |
163.096 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.28 |
-1.53 |
-14.08 |
5 |
7 |
0 |
120 |
167.128 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
3.49 |
-22.11 |
4 |
5 |
0 |
83 |
167.197 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.14 |
3.08 |
-52.15 |
3 |
5 |
-1 |
80 |
166.189 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.06 |
-3.4 |
-48.41 |
3 |
6 |
-1 |
112 |
131.067 |
1 |
↓
|
|
|
|
Analogs
-
4298371
-
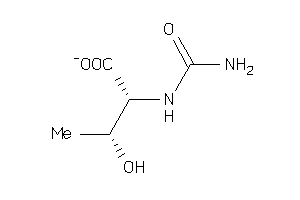
-
4298369
-
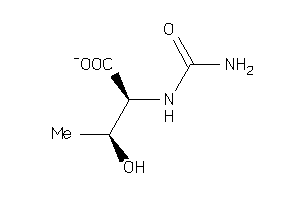
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.12 |
-6.66 |
-45.68 |
4 |
6 |
-1 |
115 |
161.137 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.48 |
-6.81 |
-56.9 |
4 |
6 |
-1 |
115 |
147.11 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.35 |
-9.58 |
-50.57 |
6 |
8 |
-1 |
150 |
175.124 |
3 |
↓
|
|
|
|
Analogs
-
2093008
-
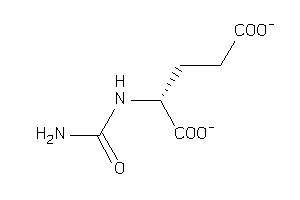
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.06 |
0.27 |
-108.25 |
3 |
7 |
-2 |
135 |
188.139 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-3.06 |
-1.71 |
-55.94 |
4 |
7 |
-1 |
133 |
189.147 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.70 |
-2.9 |
-16.68 |
4 |
4 |
0 |
86 |
114.104 |
2 |
↓
|
|
|
|
Analogs
-
39333703
-
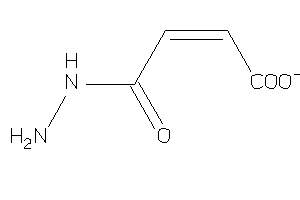
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.19 |
0.56 |
-59.21 |
2 |
4 |
-1 |
83 |
114.08 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
6219531
-
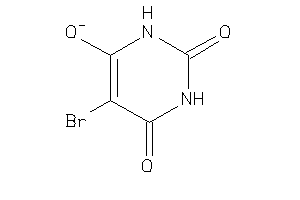
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.77 |
-4.56 |
-32.2 |
2 |
5 |
-1 |
89 |
127.079 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.41 |
-1.24 |
-35.34 |
3 |
6 |
0 |
105 |
128.091 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.41 |
-1.18 |
-35 |
3 |
6 |
0 |
105 |
128.091 |
1 |
↓
|
|
|
|
Analogs
-
32212687
-
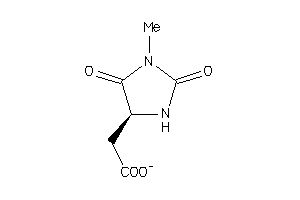
-
32212690
-
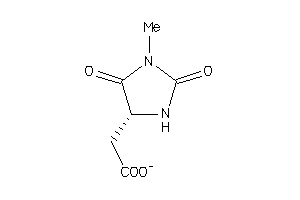
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.57 |
-4.44 |
-40.47 |
2 |
6 |
-1 |
98 |
157.105 |
2 |
↓
|
|
|
|
Analogs
-
32212687
-

-
32212690
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.57 |
-4.41 |
-40.47 |
2 |
6 |
-1 |
98 |
157.105 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
5.41 |
-37.54 |
6 |
5 |
1 |
92 |
176.203 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.64 |
-0.24 |
-33.23 |
3 |
3 |
1 |
52 |
84.102 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.64 |
-0.71 |
-7.07 |
2 |
3 |
0 |
51 |
83.094 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.28 |
1.07 |
-28.7 |
3 |
2 |
1 |
40 |
95.125 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.28 |
0.59 |
-4.62 |
2 |
2 |
0 |
39 |
94.117 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.35 |
0.7 |
-26 |
5 |
3 |
1 |
66 |
110.14 |
0 |
↓
|
|
|
|
Analogs
-
32192252
-
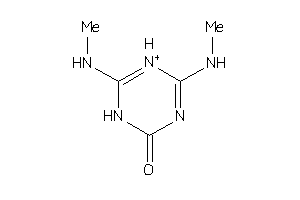
-
2382751
-
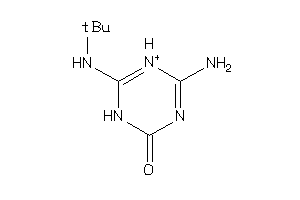
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.28 |
1.87 |
-41.96 |
5 |
6 |
1 |
98 |
156.169 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.28 |
1.94 |
-40.78 |
5 |
6 |
1 |
98 |
156.169 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.28 |
1.45 |
-17.75 |
4 |
6 |
0 |
97 |
155.161 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.61 |
-1.97 |
-42.13 |
2 |
5 |
-1 |
88 |
145.529 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.60 |
-1.24 |
-41.83 |
5 |
6 |
1 |
106 |
129.099 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.45 |
0.03 |
-46.45 |
6 |
6 |
1 |
112 |
128.115 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.45 |
-0.47 |
-15.18 |
5 |
6 |
0 |
111 |
127.107 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
-1.1 |
-55.36 |
1 |
3 |
-1 |
52 |
127.148 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.63 |
-4.93 |
-6.78 |
1 |
3 |
0 |
46 |
128.156 |
0 |
↓
|
|
|
|
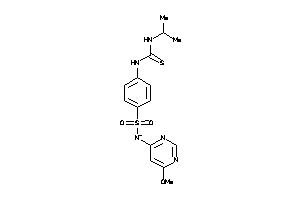
Analogs
-
6327746
-
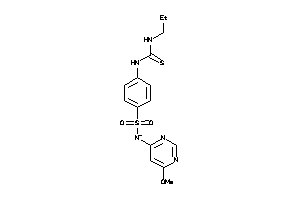
-
6361947
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
-0.13 |
-47.69 |
2 |
7 |
-1 |
109 |
279.301 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.38 |
0.31 |
-11.85 |
3 |
7 |
0 |
107 |
280.309 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.18 |
-4.52 |
-42.49 |
3 |
6 |
-1 |
111 |
154.105 |
1 |
↓
|
|