|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
9200 |
1.01 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
5500 |
1.05 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2700 |
1.11 |
Binding ≤ 10μM
|
|
CAH4-8-E |
Carbonic Anhydrase IV (cluster #8 Of 16), Eukaryotic |
Eukaryotes |
9500 |
1.00 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
8800 |
1.01 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
1.63 |
-3.7 |
1 |
1 |
0 |
20 |
94.113 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.70 |
2.74 |
-3.95 |
0 |
1 |
0 |
13 |
79.102 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.70 |
3.2 |
-29.69 |
1 |
1 |
1 |
14 |
80.11 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
4.49 |
-1.8 |
0 |
0 |
0 |
0 |
78.114 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.27 |
1.02 |
-7.62 |
0 |
2 |
0 |
20 |
73.095 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
2.39 |
-3.69 |
1 |
1 |
0 |
20 |
108.14 |
0 |
↓
|
|
|
|
Analogs
-
3881412
-
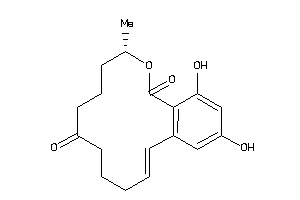
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
5.38 |
-11.84 |
2 |
5 |
0 |
84 |
318.369 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.41 |
6.28 |
-51.62 |
1 |
5 |
-1 |
87 |
317.361 |
0 |
↓
|
|
|
|
Analogs
-
3898789
-
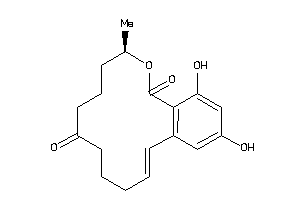
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
5.37 |
-11.72 |
2 |
5 |
0 |
84 |
318.369 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.41 |
6.33 |
-49.66 |
1 |
5 |
-1 |
87 |
317.361 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.82 |
8.43 |
-80.42 |
0 |
2 |
2 |
8 |
186.258 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
-3.99 |
-15.76 |
1 |
10 |
0 |
124 |
429.817 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
9.46 |
-14.45 |
0 |
3 |
0 |
28 |
278.335 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.18 |
2.65 |
-10.61 |
0 |
3 |
0 |
36 |
128.127 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
0.94 |
-12.33 |
3 |
6 |
0 |
96 |
290.271 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.74 |
2.04 |
-57.58 |
2 |
6 |
-1 |
99 |
289.263 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
0.94 |
-12.66 |
3 |
6 |
0 |
96 |
290.271 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.74 |
2.04 |
-57.55 |
2 |
6 |
-1 |
99 |
289.263 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
5 |
1.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
5.16 |
-1.88 |
0 |
0 |
0 |
0 |
92.141 |
0 |
↓
|
|
|
|
Analogs
-
34447578
-
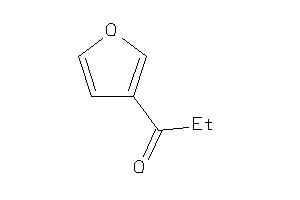
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
4.48 |
-8.94 |
0 |
3 |
0 |
47 |
166.176 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
34567204
-
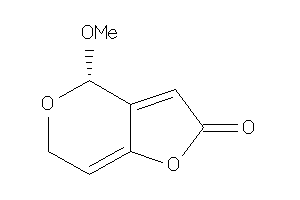
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3800 |
0.69 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
3800 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
0.94 |
-12.15 |
1 |
4 |
0 |
60 |
154.121 |
0 |
↓
|
|
|
|
Analogs
-
34567204
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3800 |
0.69 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
3800 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
0.94 |
-12.15 |
1 |
4 |
0 |
60 |
154.121 |
0 |
↓
|
|
|
|
Analogs
-
3947513
-
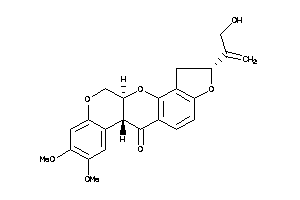
-
3947514
-
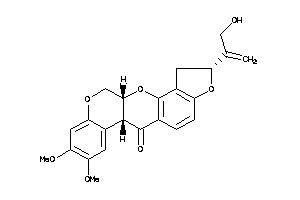
-
3947515
-
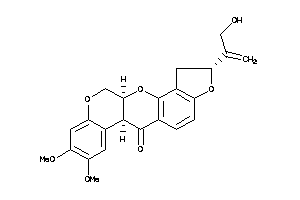
-
3947516
-
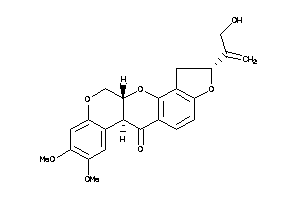
-
3978464
-
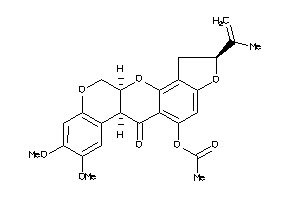
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUOX2-2-E |
Thyroid Oxidase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
NDUA4-2-E |
NADH-ubiquinone Oxidoreductase MLRQ Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUA6-2-E |
NADH-ubiquinone Oxidoreductase B14 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUA8-2-E |
NADH-ubiquinone Oxidoreductase 19 KDa Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUAA-2-E |
NADH-ubiquinone Oxidoreductase 42 KDa Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUAB-2-E |
NADH-ubiquinone Oxidoreductase Subunit B14.7 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUB5-2-E |
NADH-ubiquinone Oxidoreductase SGDH Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUB6-2-E |
NADH-ubiquinone Oxidoreductase B17 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUB7-2-E |
NADH-ubiquinone Oxidoreductase B18 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUS2-2-E |
NADH-ubiquinone Oxidoreductase 49 KDa Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUS8-2-E |
NADH-ubiquinone Oxidoreductase 23 KDa Subunit, (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NU1M-1-E |
NADH-ubiquinone Oxidoreductase Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU2M-1-E |
NADH-ubiquinone Oxidoreductase Chain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU3M-1-E |
NADH-ubiquinone Oxidoreductase Chain 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU4M-1-E |
NADH-ubiquinone Oxidoreductase Chain 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM |
|
NU5M-1-E |
NADH-ubiquinone Oxidoreductase Chain 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU6M-1-E |
NADH-ubiquinone Oxidoreductase Chain 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
Z101775-1-O |
Drosophila (cluster #1 Of 1), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50531-3-O |
Caenorhabditis Elegans (cluster #3 Of 3), Other |
Other |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z50531-1-O |
Caenorhabditis Elegans (cluster #1 Of 1), Other |
Other |
2000 |
0.28 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NDUA8_HUMAN |
P51970
|
NADH-ubiquinone Oxidoreductase 19 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUS8_HUMAN |
O00217
|
NADH-ubiquinone Oxidoreductase 23 KDa Subunit,, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUAA_HUMAN |
O95299
|
NADH-ubiquinone Oxidoreductase 42 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUS2_HUMAN |
O75306
|
NADH-ubiquinone Oxidoreductase 49 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUA6_HUMAN |
P56556
|
NADH-ubiquinone Oxidoreductase B14 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUB6_HUMAN |
O95139
|
NADH-ubiquinone Oxidoreductase B17 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUB7_HUMAN |
P17568
|
NADH-ubiquinone Oxidoreductase B18 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NU1M_HUMAN |
P03886
|
NADH-ubiquinone Oxidoreductase Chain 1, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU2M_HUMAN |
P03891
|
NADH-ubiquinone Oxidoreductase Chain 2, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU3M_HUMAN |
P03897
|
NADH-ubiquinone Oxidoreductase Chain 3, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU4M_HUMAN |
P03905
|
NADH-ubiquinone Oxidoreductase Chain 4, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU5M_HUMAN |
P03915
|
NADH-ubiquinone Oxidoreductase Chain 5, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU6M_HUMAN |
P03923
|
NADH-ubiquinone Oxidoreductase Chain 6, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NDUA4_HUMAN |
O00483
|
NADH-ubiquinone Oxidoreductase MLRQ Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUB5_HUMAN |
O43674
|
NADH-ubiquinone Oxidoreductase SGDH Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUAB_HUMAN |
Q86Y39
|
NADH-ubiquinone Oxidoreductase Subunit B14.7, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
DUOX2_HUMAN |
Q9NRD8
|
Thyroid Oxidase 2, Human |
5.1 |
0.40 |
Binding ≤ 1μM
|
|
NDUA8_HUMAN |
P51970
|
NADH-ubiquinone Oxidoreductase 19 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUS8_HUMAN |
O00217
|
NADH-ubiquinone Oxidoreductase 23 KDa Subunit,, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUAA_HUMAN |
O95299
|
NADH-ubiquinone Oxidoreductase 42 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUS2_HUMAN |
O75306
|
NADH-ubiquinone Oxidoreductase 49 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUA6_HUMAN |
P56556
|
NADH-ubiquinone Oxidoreductase B14 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUB6_HUMAN |
O95139
|
NADH-ubiquinone Oxidoreductase B17 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUB7_HUMAN |
P17568
|
NADH-ubiquinone Oxidoreductase B18 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NU1M_HUMAN |
P03886
|
NADH-ubiquinone Oxidoreductase Chain 1, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU2M_HUMAN |
P03891
|
NADH-ubiquinone Oxidoreductase Chain 2, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU3M_HUMAN |
P03897
|
NADH-ubiquinone Oxidoreductase Chain 3, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU4M_HUMAN |
P03905
|
NADH-ubiquinone Oxidoreductase Chain 4, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU5M_HUMAN |
P03915
|
NADH-ubiquinone Oxidoreductase Chain 5, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU6M_HUMAN |
P03923
|
NADH-ubiquinone Oxidoreductase Chain 6, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NDUA4_HUMAN |
O00483
|
NADH-ubiquinone Oxidoreductase MLRQ Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUB5_HUMAN |
O43674
|
NADH-ubiquinone Oxidoreductase SGDH Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUAB_HUMAN |
Q86Y39
|
NADH-ubiquinone Oxidoreductase Subunit B14.7, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
DUOX2_HUMAN |
Q9NRD8
|
Thyroid Oxidase 2, Human |
5.1 |
0.40 |
Binding ≤ 10μM
|
|
Z50531 |
Z50531
|
Caenorhabditis Elegans |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z101775 |
Z101775
|
Drosophila |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50531 |
Z50531
|
Caenorhabditis Elegans |
2000 |
0.28 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
8.74 |
-11.43 |
0 |
6 |
0 |
63 |
394.423 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
8.03 |
-12.44 |
0 |
6 |
0 |
69 |
354.337 |
2 |
↓
|
|
|
|
Analogs
-
6091129
-
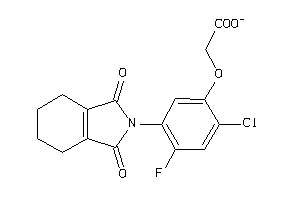
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
12.37 |
-12.72 |
0 |
6 |
0 |
75 |
423.868 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
5.9 |
-14.76 |
1 |
5 |
0 |
68 |
288.347 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.89 |
6.89 |
-26.79 |
2 |
5 |
1 |
69 |
289.355 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
5.69 |
-23.36 |
1 |
5 |
0 |
68 |
288.347 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.89 |
6.72 |
-31.09 |
2 |
5 |
1 |
69 |
289.355 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
0.78 |
-20.13 |
0 |
6 |
0 |
74 |
314.293 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
0.29 |
-21.27 |
0 |
6 |
0 |
74 |
314.293 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
0.31 |
-20.97 |
0 |
6 |
0 |
74 |
314.293 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
0.77 |
-20.13 |
0 |
6 |
0 |
74 |
314.293 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.79 |
-13.12 |
3 |
7 |
0 |
109 |
382.453 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.67 |
-16.32 |
3 |
7 |
0 |
109 |
382.453 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.27 |
-12.51 |
3 |
7 |
0 |
109 |
382.453 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
2.14 |
-14.45 |
3 |
7 |
0 |
109 |
382.453 |
5 |
↓
|
|
|
|
Analogs
-
402670
-
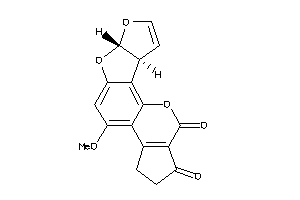
-
402671
-
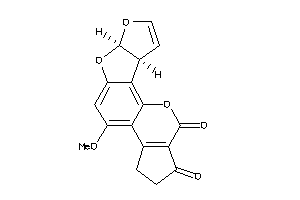
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
5.25 |
-26.99 |
1 |
6 |
0 |
86 |
298.25 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.20 |
5.61 |
-39.04 |
0 |
6 |
-1 |
89 |
297.242 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
-4.12 |
-12.79 |
4 |
6 |
0 |
103 |
298.335 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
-4.23 |
-16.59 |
4 |
6 |
0 |
103 |
298.335 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
-4.61 |
-14.27 |
4 |
6 |
0 |
103 |
298.335 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
-3.18 |
-14.13 |
4 |
6 |
0 |
103 |
298.335 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
0.53 |
-8.15 |
1 |
3 |
0 |
47 |
144.17 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
0.59 |
-8.42 |
1 |
3 |
0 |
47 |
144.17 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
0.67 |
-6.71 |
1 |
3 |
0 |
47 |
144.17 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
0.59 |
-7.48 |
1 |
3 |
0 |
47 |
144.17 |
1 |
↓
|
|
|
|
|