|
|
Analogs
-
39241039
-
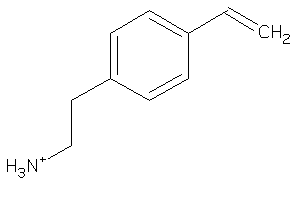
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4380 |
0.75 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.76 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5300 |
0.74 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
3.99 |
-44.19 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
890 |
0.77 |
ADME/T ≤ 10μM
|
|
CP2C9-3-E |
Cytochrome P450 2C9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6300 |
0.66 |
ADME/T ≤ 10μM
|
|
Z104283-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #2 Of 4), Other |
Other |
2285 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.69 |
4.13 |
-43.52 |
2 |
2 |
1 |
29 |
147.201 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.69 |
4.41 |
-86.78 |
3 |
2 |
2 |
31 |
148.209 |
1 |
↓
|
|
|
|
Analogs
-
4284443
-
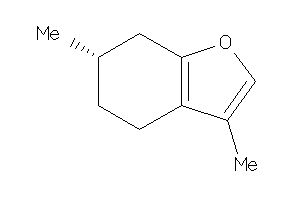
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-2-E |
Cytochrome P450 2A6 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
840 |
0.77 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
6.42 |
-3.49 |
0 |
1 |
0 |
13 |
150.221 |
0 |
↓
|
|
|
|
Analogs
-
4098388
-
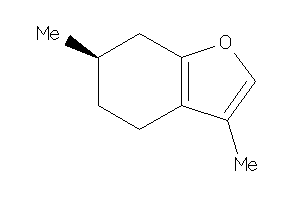
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-2-E |
Cytochrome P450 2A6 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
840 |
0.77 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
6.42 |
-3.49 |
0 |
1 |
0 |
13 |
150.221 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 46 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3280 |
0.96 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3920 |
0.95 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.73 |
4.22 |
-6.49 |
0 |
1 |
0 |
17 |
106.124 |
1 |
↓
|
|
|
|
Analogs
-
34426560
-
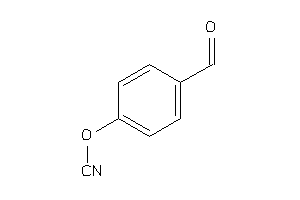
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5230 |
0.74 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5150 |
0.74 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
3.53 |
-7.83 |
0 |
2 |
0 |
26 |
136.15 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NFKB1-1-E |
Nuclear Factor NF-kappa-B P105 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.55 |
Binding ≤ 10μM
|
|
CP2A6-3-E |
Cytochrome P450 2A6 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
6.3 |
-12.66 |
0 |
3 |
0 |
43 |
186.166 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.80 |
ADME/T ≤ 10μM
|
|
CP2C9-3-E |
Cytochrome P450 2C9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5900 |
0.56 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5700 |
0.56 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
2.65 |
-48.73 |
3 |
3 |
1 |
54 |
175.211 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.23 |
2.93 |
-89.93 |
4 |
3 |
2 |
55 |
176.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1500 |
0.63 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
6.65 |
-3.2 |
0 |
1 |
0 |
13 |
254.152 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.78 |
6.93 |
-32.43 |
1 |
1 |
1 |
14 |
255.16 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
710 |
0.61 |
ADME/T ≤ 10μM
|
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
9800 |
0.50 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
3.38 |
-10.44 |
1 |
4 |
0 |
59 |
188.186 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.46 |
3.66 |
-40.55 |
2 |
4 |
1 |
60 |
189.194 |
2 |
↓
|
|
|
|
Analogs
-
34427699
-
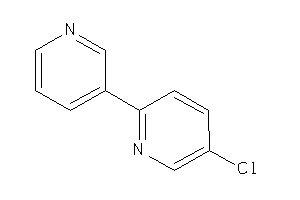
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7700 |
0.60 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
4.59 |
-6.58 |
0 |
2 |
0 |
26 |
156.188 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.52 |
4.82 |
-33.75 |
1 |
2 |
1 |
27 |
157.196 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
4500 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
5.7 |
-3.77 |
0 |
1 |
0 |
13 |
240.125 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.21 |
5.99 |
-34.13 |
1 |
1 |
1 |
14 |
241.133 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.59 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.85 |
4.19 |
-49.02 |
3 |
2 |
1 |
41 |
185.25 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.85 |
4.48 |
-86.48 |
4 |
2 |
2 |
42 |
186.258 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
790 |
0.66 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
0.23 |
-7.5 |
0 |
2 |
0 |
30 |
189.239 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
620 |
0.79 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.41 |
5.12 |
-7.99 |
0 |
3 |
0 |
31 |
145.165 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.41 |
5.62 |
-35.8 |
1 |
3 |
1 |
32 |
146.173 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.41 |
5.41 |
-41.81 |
1 |
3 |
1 |
32 |
146.173 |
1 |
↓
|
|
|
|
Analogs
-
34427699
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2700 |
0.60 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
5.22 |
-6.4 |
0 |
2 |
0 |
26 |
170.215 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.97 |
5.45 |
-32.04 |
1 |
2 |
1 |
27 |
171.223 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.65 |
ADME/T ≤ 10μM
|
|
CP2E1-2-E |
Cytochrome P450 2E1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.69 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
0.15 |
-4.63 |
0 |
1 |
0 |
13 |
161.229 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.44 |
0.25 |
-33.68 |
1 |
1 |
1 |
14 |
162.237 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6600 |
0.60 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
4.37 |
-6.47 |
0 |
2 |
0 |
26 |
156.188 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.59 |
4.83 |
-37.7 |
1 |
2 |
1 |
27 |
157.196 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5600 |
0.57 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
2.91 |
-7.1 |
1 |
2 |
0 |
33 |
191.255 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.02 |
3.18 |
-36.84 |
2 |
2 |
1 |
34 |
192.263 |
2 |
↓
|
|
|
|
Analogs
-
1729595
-
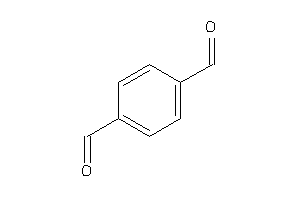
Draw
Identity
99%
90%
80%
70%
Vendors
And 59 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4680 |
0.83 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4880 |
0.83 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
1.93 |
-6.42 |
0 |
1 |
0 |
17 |
120.151 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-2-E |
Cytochrome P450 1A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2300 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5500 |
0.61 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2500 |
0.65 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.61 |
-2.8 |
0 |
0 |
0 |
0 |
197.064 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7300 |
0.65 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
670 |
0.79 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
6.84 |
-3.85 |
0 |
0 |
0 |
0 |
146.164 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.71 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
550 |
0.80 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.37 |
-3.15 |
0 |
0 |
0 |
0 |
207.07 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 22 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.67 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
1.96 |
-3.58 |
0 |
0 |
0 |
0 |
156.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.68 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1700 |
0.67 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.99 |
8.08 |
-3.61 |
0 |
0 |
0 |
0 |
156.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
2500 |
0.65 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
7.61 |
-2.94 |
0 |
0 |
0 |
0 |
197.064 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7000 |
0.66 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2400 |
0.72 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
7.42 |
-3.56 |
0 |
0 |
0 |
0 |
142.201 |
0 |
↓
|
|
|
|
Analogs
-
34975325
-
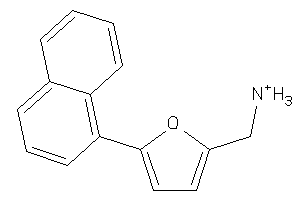
-
34975333
-
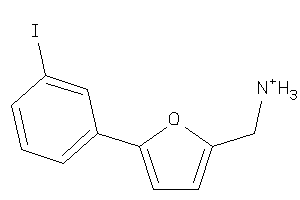
-
36984526
-
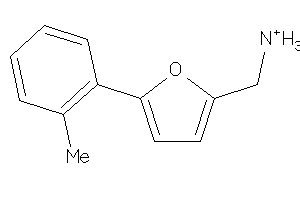
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.67 |
ADME/T ≤ 10μM
|
|
CP2B6-3-E |
Cytochrome P450 2B6 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3800 |
0.58 |
ADME/T ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
900 |
0.65 |
ADME/T ≤ 10μM
|
|
CP2E1-2-E |
Cytochrome P450 2E1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.66 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
4.42 |
-45.89 |
3 |
2 |
1 |
41 |
174.223 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
400 |
0.69 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
1.9 |
-54.15 |
3 |
4 |
1 |
67 |
176.199 |
2 |
↓
|
|
|
|
Analogs
-
4365454
-
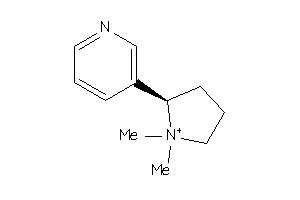
-
1798
-
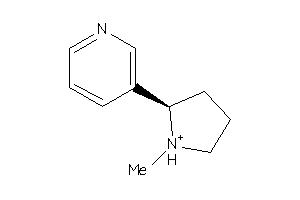
Draw
Identity
99%
90%
80%
70%
Vendors
And 41 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.94 |
Binding ≤ 10μM |
|
ACHA-4-E |
Acetylcholine Receptor Protein Alpha Chain (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM
|
|
ACHA2-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.92 |
Binding ≤ 10μM
|
|
ACHA3-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
47 |
0.85 |
Binding ≤ 10μM
|
|
ACHA4-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.61 |
Binding ≤ 10μM
|
|
ACHA5-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM
|
|
ACHA6-4-E |
Neuronal Acetylcholine Receptor Subunit Alpha-6 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM
|
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
770 |
0.71 |
Binding ≤ 10μM
|
|
ACHA9-4-E |
Neuronal Acetylcholine Receptor Protein Alpha-9 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM
|
|
ACHB-1-E |
Acetylcholine Receptor Protein Beta Chain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.95 |
Binding ≤ 10μM
|
|
ACHB2-4-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
5500 |
0.61 |
Binding ≤ 10μM
|
|
ACHB3-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-3 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.66 |
Binding ≤ 10μM
|
|
ACHB4-5-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.62 |
Binding ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6300 |
0.61 |
Binding ≤ 10μM
|
|
ACHG-1-E |
Acetylcholine Receptor Protein Gamma Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6300 |
0.61 |
Binding ≤ 10μM
|
|
ACHP-2-E |
Acetylcholine-binding Protein (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
63 |
0.84 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3610 |
0.63 |
Binding ≤ 10μM
|
|
Q8BGP7-1-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.99 |
Binding ≤ 10μM
|
|
ACHA3-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9400 |
0.59 |
Functional ≤ 10μM
|
|
ACHA4-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6600 |
0.60 |
Functional ≤ 10μM
|
|
ACHA7-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.68 |
Functional ≤ 10μM
|
|
ACHD-1-E |
Acetylcholine Receptor Protein Delta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7100 |
0.60 |
Functional ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4400 |
0.62 |
ADME/T ≤ 10μM
|
|
Z104281-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #2 Of 2), Other |
Other |
47 |
0.85 |
Binding ≤ 10μM
|
|
Z104282-1-O |
Acetylcholine Receptor; Alpha1/beta1/delta/gamma (cluster #1 Of 1), Other |
Other |
6270 |
0.61 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
9 |
0.94 |
Binding ≤ 10μM
|
|
Z104284-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta4 (cluster #1 Of 1), Other |
Other |
70 |
0.83 |
Binding ≤ 10μM |
|
Z104285-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 1), Other |
Other |
129 |
0.80 |
Binding ≤ 10μM
|
|
Z104286-2-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #2 Of 2), Other |
Other |
6 |
0.96 |
Binding ≤ 10μM |
|
Z104287-3-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #3 Of 3), Other |
Other |
530 |
0.73 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
530 |
0.73 |
Binding ≤ 10μM
|
|
Z104289-2-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #2 Of 2), Other |
Other |
46 |
0.86 |
Binding ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
10 |
0.93 |
Binding ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 1), Other |
Other |
1 |
1.05 |
Binding ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 2), Other |
Other |
9400 |
0.59 |
Functional ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 1), Other |
Other |
8000 |
0.59 |
Functional ≤ 10μM
|
|
Z104290-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 1), Other |
Other |
7 |
0.95 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
40 |
0.86 |
Functional ≤ 10μM
|
|
Z81074-1-O |
K177cell Line (cluster #1 Of 1), Other |
Other |
5000 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
5.14 |
-35.14 |
1 |
2 |
1 |
17 |
163.244 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
80 |
0.76 |
ADME/T ≤ 10μM
|
|
CP2C9-3-E |
Cytochrome P450 2C9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8900 |
0.54 |
ADME/T ≤ 10μM
|
|
CP2CJ-1-E |
Cytochrome P450 2C19 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.61 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
3.65 |
-48.44 |
3 |
2 |
1 |
41 |
191.279 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.87 |
3.92 |
-87.82 |
4 |
2 |
2 |
42 |
192.287 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
90 |
0.99 |
ADME/T ≤ 10μM
|
|
CP2C9-3-E |
Cytochrome P450 2C9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1200 |
0.83 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.29 |
2.25 |
-48.3 |
3 |
2 |
1 |
41 |
133.174 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.29 |
2.53 |
-92.62 |
4 |
2 |
2 |
42 |
134.182 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
700 |
0.78 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
5.11 |
-5.23 |
0 |
1 |
0 |
13 |
161.229 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.25 |
5.4 |
-33.49 |
1 |
1 |
1 |
14 |
162.237 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
8600 |
0.51 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
6.03 |
-7.55 |
0 |
2 |
0 |
30 |
203.266 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.58 |
6.3 |
-42.28 |
1 |
2 |
1 |
31 |
204.274 |
2 |
↓
|
|
|
|
Analogs
-
2392664
-
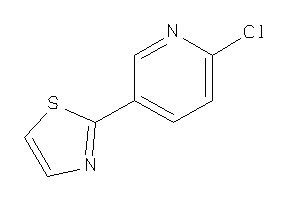
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3200 |
0.70 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
3.65 |
-6.72 |
0 |
2 |
0 |
26 |
162.217 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
600 |
0.62 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
5.53 |
-42.7 |
2 |
2 |
1 |
29 |
205.306 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.24 |
5.8 |
-82.82 |
3 |
2 |
2 |
31 |
206.314 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP1A2-2-E |
Cytochrome P450 1A2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.60 |
ADME/T ≤ 10μM
|
|
CP2A5-2-E |
Cytochrome P450 2A5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9908 |
0.58 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8300 |
0.59 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
7.95 |
-3.78 |
0 |
0 |
0 |
0 |
156.228 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-5-E |
Cytochrome P450 2A6 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
5400 |
0.67 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.80 |
7.27 |
-3.23 |
0 |
0 |
0 |
0 |
162.619 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
300 |
0.70 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
0.65 |
-46.16 |
4 |
4 |
1 |
69 |
175.215 |
2 |
↓
|
|
|
|
Analogs
-
34439407
-
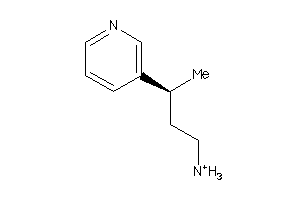
-
34439408
-
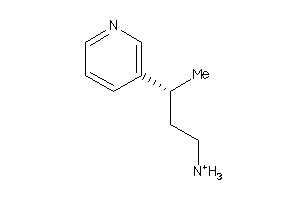
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6600 |
0.73 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
2.3 |
-46.27 |
3 |
2 |
1 |
41 |
137.206 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.20 |
2.58 |
-89.9 |
4 |
2 |
2 |
42 |
138.214 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3300 |
0.70 |
ADME/T ≤ 10μM
|
|
CP2E1-2-E |
Cytochrome P450 2E1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9700 |
0.64 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
1.14 |
-3.46 |
0 |
0 |
0 |
0 |
160.241 |
1 |
↓
|
|
|
|
Analogs
-
39250338
-
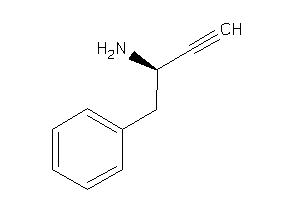
-
39250339
-
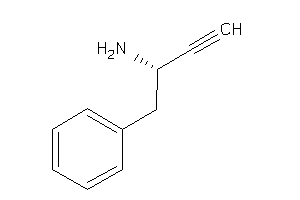
Draw
Identity
99%
90%
80%
70%
Vendors
-
- API0000117
- CCH0003609
- API0002279
-
-
And 12 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4.01 |
-39.14 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
Analogs
-
39250338
-

-
39250339
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-1-E |
Serotonin 1a (5-HT1a) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1D-1-E |
Serotonin 1d (5-HT1d) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7660 |
0.72 |
Binding ≤ 10μM
|
|
SC6A3-2-E |
Dopamine Transporter (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
920 |
0.85 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2850 |
0.78 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3500 |
0.76 |
ADME/T ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
490 |
0.88 |
Functional ≤ 10μM
|
|
Z50597-12-O |
Rattus Norvegicus (cluster #12 Of 12), Other |
Other |
800 |
0.85 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4 |
-39.2 |
3 |
1 |
1 |
28 |
136.218 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP2A6-4-E |
Cytochrome P450 2A6 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
4100 |
0.63 |
ADME/T ≤ 10μM
|
|
CP2E1-2-E |
Cytochrome P450 2E1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9600 |
0.59 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.94 |
4.27 |
-6.81 |
0 |
2 |
0 |
26 |
176.244 |
1 |
↓
|
|
|
|
Analogs
-
39291087
-
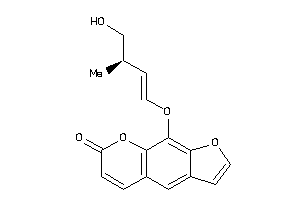
-
39291089
-
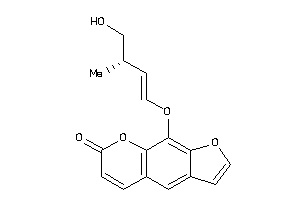
Draw
Identity
99%
90%
80%
70%
Vendors
And 47 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
6.1 |
-16.38 |
0 |
4 |
0 |
53 |
216.192 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 60 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TAAR1-2-E |
Trace Amine-associated Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
129 |
1.07 |
Functional ≤ 10μM
|
|
CP2A5-1-E |
Cytochrome P450 2A5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.86 |
ADME/T ≤ 10μM
|
|
CP2A6-1-E |
Cytochrome P450 2A6 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4410 |
0.83 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.92 |
3.31 |
-44.08 |
3 |
1 |
1 |
28 |
122.191 |
2 |
↓
|
|